#2022/04/20 追記
FiltlongはONTのロングリードのクオリティ分析やクオリティ、リード長のトリミングが行えるツール。ウルトラロングリードを低クオリティ領域でカットして、分割出力する機能も備える。2018年4月現在Githubで公開されている。
インストール
mac os10.13に導入した。
依存
https://github.com/rrwick/Filtlong
#conda
mamba install -c bioconda filtlong -y
git clone https://github.com/rrwick/Filtlong.git
cd Filtlong
make -j 4
bin/filtlong -h
usage: filtlong {OPTIONS} [input_reads]
Filtlong: a quality filtering tool for Nanopore and PacBio reads
positional arguments:
input_reads input long reads to be filtered
optional arguments:
output thresholds:
-t[int], --target_bases [int] keep only the best reads up to this many total bases
-p[float], --keep_percent [float] keep only this percentage of the best reads (measured by bases)
--min_length [int] minimum length threshold
--max_length [int] maximum length threshold
--min_mean_q [float] minimum mean quality threshold
--min_window_q [float] minimum window quality threshold
external references (if provided, read quality will be determined using these instead of from the Phred scores):
-a[file], --assembly [file] reference assembly in FASTA format
-1[file], --illumina_1 [file] reference Illumina reads in FASTQ format
-2[file], --illumina_2 [file] reference Illumina reads in FASTQ format
score weights (control the relative contribution of each score to the final read score):
--length_weight [float] weight given to the length score (default: 1)
--mean_q_weight [float] weight given to the mean quality score (default: 1)
--window_q_weight [float] weight given to the window quality score (default: 1)
read manipulation:
--trim trim non-k-mer-matching bases from start/end of reads
--split [split] split reads at this many (or more) consecutive non-k-mer-matching bases
other:
--window_size [int] size of sliding window used when measuring window quality (default: 250)
--verbose verbose output to stderr with info for each read
--version display the program version and quit
-h, --help display this help menu
For more information, go to: https://github.com/rrwick/Filtlong
ラン
リファレンスがない時のクオリティ分析。
filtlong --min_length 1000 --keep_percent 90 \
--target_bases 500000000 input.fastq.gz | \
gzip - > output.fastq.gz
- --min_length 1000 Discard any read which is shorter than 1 kbp.
- --keep_percent 90 Throw out the worst 10% of reads. This is measured by bp, not by read count. So this option throws out the worst 10% of read bases.
- --target_bases 500000000 Remove the worst reads until only 500 Mbp remain, useful for very large read sets. If the input read set is less than 500 Mbp, this setting will have no effect.
- input.fastq.gz The input long reads to be filtered (must be FASTQ format).
リファレンスとなるハイクオリティなショートリードがある時のクオリティ分析。このモードではロングリードのquality scoreは使わずショートリードとk-merマッチを行う。ONTのロングリードのクオリティスコアを使わないことで分析精度が上がるとされる。
filtlong -1 illumina_1.fastq.gz -2 illumina_2.fastq.gz \
--min_length 1000 --keep_percent 90 --target_bases 500000000 \
--trim --split 500 input.fastq.gz | gzip > output.fastq.gz
- -1 illumina_1.fastq.gz -2 illumina_2.fastq.gz Use Illumina reads as an external reference. You can instead use "-a" to provide an assembly as a reference, but Illumina reads are preferable if available.
クオリティ分析とトリミングおよびウルトラロングリードのsplit triminng。
filtlong -1 illumina_1.fastq.gz -2 illumina_2.fastq.gz --min_length 1000 \
--keep_percent 90 --target_bases 500000000 --trim --split 500 \
input.fastq.gz | gzip > output.fastq.gz
- --trim Trim bases from the start and end of reads which do not match a k-mer in the reference. This ensures the each read starts and ends with good sequence.
- --split 500 Split reads whenever 500 consequence bases fail to match a k-mer in the reference. This serves to remove very poor parts of reads while keeping the good parts. A lower value will split more aggressively and a higher value will be more conservative.
split機能(--split)によりウルトラロングリードのpoor qualityな領域でリードが切断され、500bp以上の領域が確保できれば別リードとして出力される。
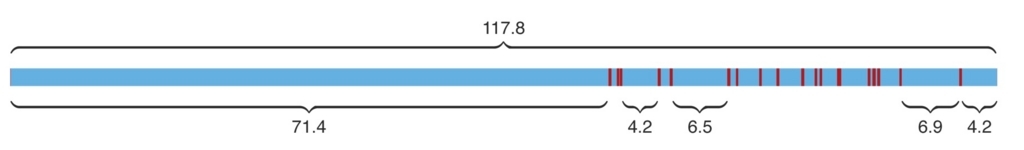
公式より
リード長優先トリミング
filtlong -1 illumina_1.fastq.gz -2 illumina_2.fastq.gz --min_length 1000\
--keep_percent 90 --target_bases 500000000 --trim --split 1000 --length_weight 10 \
input.fastq.gz | gzip > output.fastq.gz
- --length_weight 10 A length weight of 10 (instead of the default of 1) makes read length more important when choosing the best reads.
- --split 1000 This larger split value makes Filtlong less likely to split a read. I.e. a read has to have a lot of consecutive bad bases before it gets split. This helps to keep the output reads longer.
クオリティ優先トリミング
filtlong -1 illumina_1.fastq.gz -2 illumina_2.fastq.gz --min_length 1000 \
--keep_percent 90 --target_bases 500000000 --trim --split 100 --mean_q_weight 10 \
input.fastq.gz | gzip > output.fastq.gz
- --mean_q_weight 10 A mean quality weight of 10 (instead of the default of 1) makes mean read quality more important when choosing the best reads.
- --split 100 This smaller split value makes Filtlong split reads more often. I.e. even a relatively small stretch of bad bases will result in a split, giving shorter reads but of higher quality.
Githubの公開ページでは、バクテリアのmultiplex minion sequenceのデータを使った例が示されています。
引用
https://github.com/rrwick/Filtlong