2018 10/7 文章訂正、10/12 dockerコンテナを使ったランの流れ追加
2019 4/11 dockerを使ったランで表も保存するよう修正、/16 インストール追記、11/28 インストール追記、データベース作成の流れを修正、12/6 バージョンアップ追記
2021 1/15 バージョンアップ追記、7/31 追記,
2022/03/29 月qaコマンド追記、07/14 タイトル変更
ドラフトゲノムからゲノムの完全さを正確に見積もるには、ゲノムの完全さと汚染の度合いの正確な推定が必要となる。そのための方法として、一般にすべての細菌または古細菌ゲノムにわたって保存されたマーカー遺伝子を利用することができる。CheckMは、参照ゲノムツリー内のゲノムに特異的なマーカー遺伝子セット情報を使用して、ゲノムの完全性と汚染の正確な推定値を提供する。系統内で単一コピーの遺伝子を元に、ゲノムが完全であるか、コンタミがあるか分析している。
HP
http://ecogenomic.org/software
https://github.com/Ecogenomics/CheckM/wiki
2019 12/6 追記
CheckM is now Python 3: https://t.co/QAuP1q8dwu
— Donovan Parks (@donovan_parks) 2019年12月1日
インストール
依存
python関連
- python >= 2.7 and < 3.0 ==> python3
- numpy >= 1.8.0
- scipy >= 0.9.0
- matplotlib >= 1.3.1
- pysam >= 0.8.3
- dendropy >= 4.0.0
そのほか
- HMMER (>=3.1b1)
- prodigal (2.60 or >=2.6.1)
- pplacer (>=1.1)(binaryリンク)
HMMERとprodigalはbrewでインストールできる。pplacerはbinaryを上記リンクからダウンロードする。
sudo pip install numpy
sudo pip install checkm-genome #依存するpythonライブラリもインストールされる
#bioconda(link)python2.7仮想環境に入れる
conda create -n checkm -c bioconda -y checkm-genome python=2.7
conda activate checkm
#2019 12/6 追記 python3になっています
#(HP)CheckM v1.1.3 requires Python 3.
mamba create -n checkm -c bioconda -y checkm-genome python=3.8
conda activate checkm
mamba install -c bioconda -y pplacer
> checkm
$ checkm
...::: CheckM v1.0.7 :::...
Lineage-specific marker set:
tree -> Place bins in the reference genome tree
tree_qa -> Assess phylogenetic markers found in each bin
lineage_set -> Infer lineage-specific marker sets for each bin
Taxonomic-specific marker set:
taxon_list -> List available taxonomic-specific marker sets
taxon_set -> Generate taxonomic-specific marker set
Apply marker set to genome bins:
analyze -> Identify marker genes in bins
qa -> Assess bins for contamination and completeness
Common workflows (combines above commands):
lineage_wf -> Runs tree, lineage_set, analyze, qa
taxonomy_wf -> Runs taxon_set, analyze, qa
Bin QA plots:
bin_qa_plot -> Bar plot of bin completeness, contamination, and strain heterogeneity
Reference distribution plots:
gc_plot -> Create GC histogram and delta-GC plot
coding_plot -> Create coding density (CD) histogram and delta-CD plot
tetra_plot -> Create tetranucleotide distance (TD) histogram and delta-TD plot
dist_plot -> Create image with GC, CD, and TD distribution plots together
General plots:
nx_plot -> Create Nx-plots
len_plot -> Cumulative sequence length plot
len_hist -> Sequence length histogram
marker_plot -> Plot position of marker genes on sequences
par_plot -> Parallel coordinate plot of GC and coverage
gc_bias_plot -> Plot bin coverage as a function of GC
Sequence subspace plots:
cov_pca -> PCA plot of coverage profiles
tetra_pca -> PCA plot of tetranucleotide signatures
Bin exploration and modification:
unique -> Ensure no sequences are assigned to multiple bins
merge -> Identify bins with complementary sets of marker genes
bin_compare -> Compare two sets of bins (e.g., from alternative binning methods)
bin_union -> [Experimental] Merge multiple binning efforts into a single bin set
modify -> [Experimental] Modify sequences in a bin
outliers -> [Experimental] Identify outlier in bins relative to reference distributions
Utility functions:
unbinned -> Identify unbinned sequences
coverage -> Calculate coverage of sequences
tetra -> Calculate tetranucleotide signature of sequences
profile -> Calculate percentage of reads mapped to each bin
join_tables -> Join tab-separated value tables containing bin information
ssu_finder -> Identify SSU (16S/18S) rRNAs in sequences
Use: 'checkm data' to find, download and install database updates
Use: checkm <command> -h for command specific help
データベースの準備
checkmのデータベースフォルダを作成する。
checkm data setRoot ~/Document/checkm_database
この時点ではディレクトリは空になっている。依存するデータセットをダウンロードし、このディレクトリに収納する。マニュアルでダウンロードするなら
https://data.ace.uq.edu.au/public/CheckM_databases/
にアクセスして、checkm_data_2015_01_16.tar.gzを選択。ダウンロードして、先ほどのディレクトリに解凍する。
cd ~/Document/checkm_database
wget https://data.ace.uq.edu.au/public/CheckM_databases/checkm_data_2015_01_16.tar.gz
tar zxvf checkm_data_2015_01_16.tar.gz
2021 7/31
(データベースが古く、最新の公開ゲノムでマーカー遺伝子を定義し直すと結果が変わりそうだが、checkM2が現在開発中で(まだ初期段階)、更新予定はないらしい)。
テストランを行う。
checkm test ~/checkm_test_results
$ sudo checkm test ~/checkm_test
*******************************************************************************
-bash: results: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ [CheckM - Test] Processing E.coli K12-W3310 to verify operation of CheckM.
-bash: [CheckM: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ *******************************************************************************
-bash: results: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ [Step 1]: Verifying tree command.
-bash: [Step: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ *******************************************************************************
-bash: results: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ [CheckM - tree] Placing bins in reference genome tree.
-bash: [CheckM: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ *******************************************************************************
-bash: results: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Identifying marker genes in 1 bins with 1 threads:
-bash: Identifying: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished processing 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$ Saving HMM info to file.
-bash: Saving: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Calculating genome statistics for 1 bins with 1 threads:
-bash: Calculating: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished processing 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Extracting marker genes to align.
-bash: Extracting: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Parsing HMM hits to marker genes:
-bash: Parsing: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished parsing hits for 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$ Extracting 43 HMMs with 1 threads:
-bash: Extracting: command not found
Finished extracting 43 of 43 (100.00%) HMMs.
Aligning 43 marker genes with 1 threads:
Finished aligning 43 of 43 (100.00%) marker genes.
Reading marker alignment files.
Concatenating alignments.
Placing 1 bins into the genome tree with pplacer (be patient).
{ Current stage: 0:02:17.339 || Total: 0:02:17.339 }
[Passed]
[Step 2]: Verifying tree_qa command.
*******************************************************************************
[CheckM - tree_qa] Assessing phylogenetic markers found in each bin.
*******************************************************************************
Reading HMM info from file.
Parsing HMM hits to marker genes:
Finished parsing hits for 1 of 1 (100.00%) bins.
QA information written to: /home/kazu/checkm_test/results/tree_qa_test.tsv
{ Current stage: 0:00:00.334 || Total: 0:02:17.673 }
[Passed]
[Step 3]: Verifying lineage_set command.
*******************************************************************************
[CheckM - lineage_set] user-no-MacBook-Pro-2:checkm_test_results user$ Finished extracting 43 of 43 (100.00%) HMMs.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$ Aligning 43 marker genes with 1 threads:
-bash: Aligning: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished aligning 43 of 43 (100.00%) marker genes.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Reading marker alignment files.
-bash: Reading: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Concatenating alignments.
-bash: Concatenating: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Placing 1 bins into the genome tree with pplacer (be patient).
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:02:17.339 || Total: 0:02:17.339 }
>
> [Passed]
>
>
> [Step 2]: Verifying tree_qa command.
>
> *******************************************************************************
> [CheckM - tree_qa] Assessing phylogenetic markers found in each bin.
> *******************************************************************************
>
> Reading HMM info from file.
> Parsing HMM hits to marker genes:
> Finished parsing hits for 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ QA information written to: /home/kazu/checkm_test/results/tree_qa_test.tsv
-bash: QA: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:00:00.334 || Total: 0:02:17.673 }
>
> [Passed]
>
>
> [Step 3]: Verifying lineage_set command.
>
> *******************************************************************************
> [CheckM - lineage_set] Inferring lineage-specific marker sets.
> *******************************************************************************
>
> Reading HMM info from file.
> Parsing HMM hits to marker genes:
> Finished parsing hits for 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Determining marker sets for each genome bin.
-bash: Determining: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished processing 1 of 1 (100.00%) bins (current: 637000110).
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Marker set written to: /home/kazu/checkm_test/results/lineage_set_test.tsv
-bash: Marker: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:00:00.717 || Total: 0:02:18.391 }
>
> *******************************************************************************
> [CheckM - lineage_set] Inferring lineage-specific marker sets.
> *******************************************************************************
>
> Reading HMM info from file.
> Parsing HMM hits to marker genes:
> Finished parsing hits for 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Determining marker sets for each genome bin.
-bash: Determining: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished processing 1 of 1 (100.00%) bins (current: 637000110).
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ Marker set written to: /home/kazu/checkm_test/results/lineage_set_test.tsv
-bash: Marker: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:00:00.677 || Total: 0:02:19.068 }
>
> [Passed]
>
>
> [Step 4]: Verifying analyze command.
>
> *******************************************************************************
> [CheckM - analyze] Identifying marker genes in bins.
> *******************************************************************************
>
> Identifying marker genes in 1 bins with 1 threads:
> Finished processing 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$ Saving HMM info to file.
-bash: Saving: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:03:30.056 || Total: 0:05:49.125 }
>
> Parsing HMM hits to marker genes:
> Finished parsing hits for 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$ Aligning marker genes with multiple hits in a single bin:
-bash: Aligning: command not found
user-no-MacBook-Pro-2:checkm_test_results user$ Finished processing 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:00:00.854 || Total: 0:05:49.980 }
>
> Calculating genome statistics for 1 bins with 1 threads:
> Finished processing 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:00:00.189 || Total: 0:05:50.170 }
>
> [Passed]
>
>
> [Step 5]: Verifying qa command.
>
> *******************************************************************************
> [CheckM - qa] Tabulating genome statistics.
> *******************************************************************************
>
> Calculating AAI between multi-copy marker genes.
>
> Reading HMM info from file.
> Parsing HMM hits to marker genes:
> Finished parsing hits for 1 of 1 (100.00%) bins.
-bash: syntax error near unexpected token `('
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ QA information written to: /home/kazu/checkm_test/results/qa_test.tsv
-bash: QA: command not found
user-no-MacBook-Pro-2:checkm_test_results user$
user-no-MacBook-Pro-2:checkm_test_results user$ { Current stage: 0:00:00.983 || Total: 0:05:51.154 }
>
> [Passed]
>
> { Current stage: 0:00:00.007 || Total: 0:05:51.161 }
解析が終わるとホームにcheckm_test_results/ができ、その中に分析結果が出力される。
実行方法
Quick start
メタゲノムからビニングしたFASTAを収納したディレクトリを指定してランする。ビニングしたファイルの拡張子は.fnaを認識する。異なるなら"-x fa"などと指定する。標準出力のログも保存するなら1>をつける。
checkm lineage_wf -t 8 -x fna metagenome/ output 1> log
メモリが40GB以下のマシンでは、--reduced_treeをつけることでメモリ使用量を14GBまで抑えることができる。
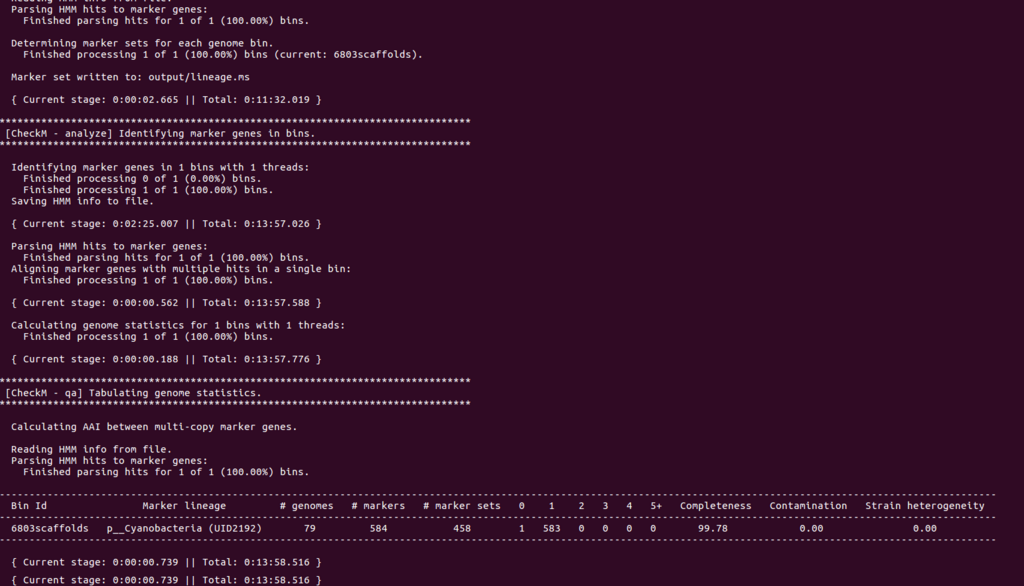
Completeness99.78、Contamination 0となった。Cyanobacteraのラベルも付いている。
純化を試みたが、まだできていないままシーケンスしたデータも解析してみる。contigは8000くらいある。
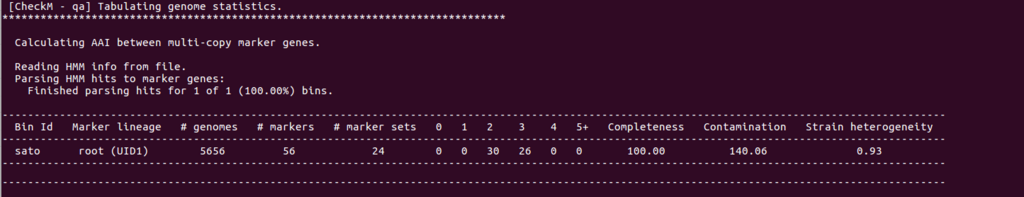
Completenessは100だが、Contamination は0でなく140となった。
wikiにはLineage-specific WorkflowとTaxonomic-specific Workflowが記載されています(リンク)。特定の系統に特異的なマーカーセットを使ってランしたり、逆に全て同じマーカーセットを使ってランする流れが載っています。興味がある人は確認して見てください。メタゲノムをビニングしたデータは持ってないので、ここではテストしません。
結果からテーブルを出力する。checkm lineage_wfの出力ディレクトリを指定する。
checkm qa chrcmM_result/lineage.ms chrcmM_result/ > result
実行方法2
dockerコンテナを利用してランするのが楽。
sstevensさんのイメージを使わせてもらう(link)。
docker pull sstevens/checkm
#例えばカレントパスをイメージの/dataと共有してrun。メモリは32g指定(*1)
docker run -m 32g -itv $PWD:/data/ sstevens/checkm
#まず
> set root w/command `checkm data setRoot /checkm-data`
#ランする
> checkm lineage_wf -t 4 -x fasta /data/metagenome_bins/ output
#ランする。表も保存する
> checkm lineage_wf -t 4 -x fasta /data/metagenome_bins/ output > table
指定ディレクトリに 複数fastaがある場合、そのfastaごとに評価される。
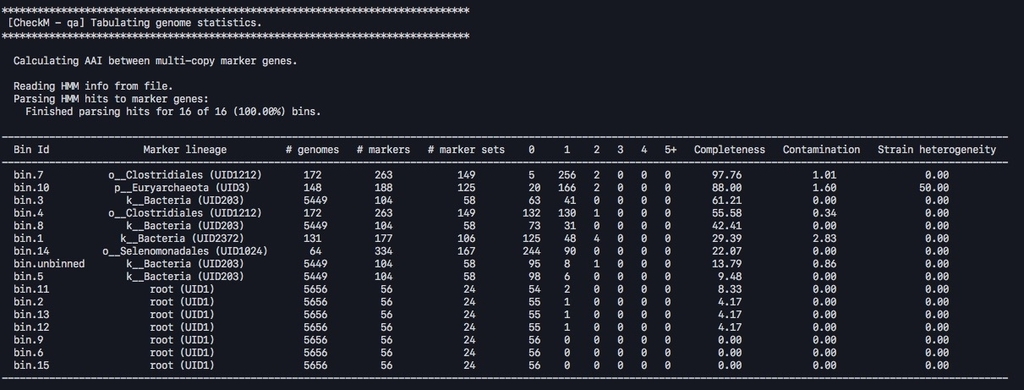
追記
Reading marker alignment files.
Concatenating alignments.
Placing 1 bins into the genome tree with pplacer (be patient).
Killed
のようなメッセージが出たら、メモリ不足の可能性があります。単純にデータを減らすか、--reduced_treeをつけてやり直してみて下さい。
追記
他にも様々なコマンドがある。
> checkm coverage
$ checkm coverage
usage: checkm coverage [-h] [-x EXTENSION] [-r] [-a MIN_ALIGN]
[-e MAX_EDIT_DIST] [-m MIN_QC] [-t THREADS] [-q]
bin_dir output_file bam_files [bam_files ...]
checkm coverage: error: too few arguments
> checkm profile
$ checkm profile
usage: checkm profile [-h] [-f FILE] [--tab_table] [-q] coverage_file
checkm profile: error: too few arguments
checkm binned_dir/ sample1coverage sample1.bam
checkm profile sample1coverage > sample1_coverage_profile
引用
CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW.
Genome Res. 2015 Jul;25(7):1043-55.
追記1
コンタミを可視化するなら、BlobToolsが使えます。
checkmも組み込まれている包括的なメタゲノム解析パイプライン
追記
たくさんのゲノム配列をまとめて評価する。
2022/05/12
MDMcleaner に切り替えることも考えて下さい。
2022/07/13
checkM2
*1
先に右上のdockerのpreferenceからメモリ使用量の上限を上げておいて下さい。