2020 2/3 インストール追記、実行例追記
2020 8/13 インストール追記
rnaQUASTはde novo transcriptomeのアセンブルパフォーマンスを比較するツール。リファレンスゲノムやtranscriptsのカタログにアセンブルした配列をアライメントし、様々な統計データをPDFで出力する。リファンレンスの遺伝子情報(gtf)がない時でも、ラン中にGeneMarkS-Tを動かし遺伝子を予測してランすることもできる。内部でRNAのアライナーを動かし(STARやTophatが必要)、カバレッジを調べることもできる。
公式サイト
http://cab.spbu.ru/software/rnaquast/
マニュアル
http://cab.spbu.ru/files/rnaquast/release1.5.0/manual.html
インストール
ubuntu18.04LTS のマシンでテストした(macosには対応していない)。
依存
- Python 2 (2.5 or higher)
- matplotlib python package
- joblib python package
- gffutils python package (needs biopython)
- NCBI BLAST+ (blastn)
- GMAP (or BLAT) aligner
optional
- STAR (or alternatively TopHat aligner + SAM tools)
- GeneMarkS-T
gffutils、matplotlib、joblibはpipでインストールする。BLATやBLASTはbrewで導入できる。GMAPを使うなら、github(リンク)からダウンロードしてビルドする。
#bioconda (link)
conda create -n rnaquast -y
conda activate rnaquast
conda install -c bioconda -y rnaquast
>rnaQUAST.py -h
$ rnaQUAST.py -h
usage: /home/kazu/anaconda3/envs/rnaquast/bin/rnaQUAST.py [-h]
[-r REFERENCE [REFERENCE ...]]
[--gtf GTF [GTF ...]]
[--gene_db GENE_DB]
[-c TRANSCRIPTS [TRANSCRIPTS ...]]
[-psl ALIGNMENT [ALIGNMENT ...]]
[-sam READS_ALIGNMENT]
[-1 LEFT_READS]
[-2 RIGHT_READS]
[-s SINGLE_READS]
[--gmap_index GMAP_INDEX]
[-o OUTPUT_DIR]
[--test] [-d]
[-t THREADS]
[-l LABELS [LABELS ...]]
[-ss]
[--min_alignment MIN_ALIGNMENT]
[--no_plots]
[--blat] [--tophat]
[--gene_mark]
[--meta]
[--lower_threshold LOWER_THRESHOLD]
[--upper_threshold UPPER_THRESHOLD]
[--disable_infer_genes]
[--disable_infer_transcripts]
[--busco_lineage BUSCO_LINEAGE]
[--prokaryote]
QUALITY ASSESSMENT FOR TRANSCRIPTOME ASSEMBLIES /home/kazu/anaconda3/envs/rnaquast/bin/rnaQUAST.py v.1.5.1
Usage:
python /home/kazu/anaconda3/envs/rnaquast/bin/rnaQUAST.py --transcripts TRANSCRIPTS --reference REFERENCE --gtf GENE_COORDINATES
optional arguments:
-h, --help show this help message and exit
Input data:
-r REFERENCE [REFERENCE ...], --reference REFERENCE [REFERENCE ...]
Single file (or several files for meta RNA) with
reference genome in FASTA format or *.txt file with
one-per-line list of FASTA files with reference
sequences
--gtf GTF [GTF ...] File with gene coordinates (or several files or *.txt
file with one-per-line list of GTF / GFF files for
meta RNA). We recommend to use files downloaded from
GENCODE or Ensembl [GTF/GFF]
--gene_db GENE_DB Path to the gene database generated by gffutils to be
used
-c TRANSCRIPTS [TRANSCRIPTS ...], --transcripts TRANSCRIPTS [TRANSCRIPTS ...]
File(s) with transcripts [FASTA]
-psl ALIGNMENT [ALIGNMENT ...], --alignment ALIGNMENT [ALIGNMENT ...]
File(s) with transcript alignments to the reference
genome [PSL]
-sam READS_ALIGNMENT, --reads_alignment READS_ALIGNMENT
File with read alignments to the reference genome
[SAM]
-1 LEFT_READS, --left_reads LEFT_READS
File with forward paired-end reads [FASTQ or gzip-
compressed]
-2 RIGHT_READS, --right_reads RIGHT_READS
File with reverse paired-end reads [FASTQ or gzip-
compressed]
-s SINGLE_READS, --single_reads SINGLE_READS
File with unpaired reads [FASTQ or gzip-compressed]
--gmap_index GMAP_INDEX
Folder containing GMAP index for the reference genome
Basic options:
-o OUTPUT_DIR, --output_dir OUTPUT_DIR
Directory to store all results [default:
rnaQUAST_results/results_<datetime>]
--test Run rnaQUAST on the test data from the test_data
folder, output directory is rnaOUAST_test_output
-d, --debug Report detailed information, typically used only for
detecting problems.
Advanced options:
-t THREADS, --threads THREADS
Maximum number of threads, default: min(number of CPUs
/ 2, 16)
-l LABELS [LABELS ...], --labels LABELS [LABELS ...]
Name(s) of assemblies that will be used in the reports
-ss, --strand_specific
Set if transcripts were assembled using strand-
specific RNA-Seq data
--min_alignment MIN_ALIGNMENT
Minimal alignment length, default: 50
--no_plots Do not draw plots (to speed up computation)
--blat Run with BLAT alignment tool
(http://hgwdev.cse.ucsc.edu/~kent/exe/) instead of
GMAP
--tophat Run with TopHat tool
(https://ccb.jhu.edu/software/tophat/index.shtml)
instead of STAR
--gene_mark Run with GeneMarkS-T tool
(http://topaz.gatech.edu/GeneMark/)
--meta Run QUALITY ASSESSMENT FOR METATRANSCRIPTOME
ASSEMBLIES
--lower_threshold LOWER_THRESHOLD
Lower threshold for x-assembled/covered/matched
metrics, default: 0.5
--upper_threshold UPPER_THRESHOLD
Upper threshold for x-assembled/covered/matched
metrics, default: 0.95
--prokaryote Use this option if the genome is prokaryotic
Gffutils related options:
--disable_infer_genes
Use this option if your GTF file already contains
genes records
--disable_infer_transcripts
Use this option if your GTF already contains
transcripts records
BUSCO related options:
Run with BUSCO tool (http://busco.ezlab.org/). Path to
the BUSCO lineage data to be used (Eukaryota, Metazoa,
Arthropoda, Vertebrata or Fungi)
Don't forget to add GMAP (or BLAT) to PATH.
(rnaquast) kazu@kazu:~/Downloads$
ラン
アセンブルしたfastaとリファレンスのゲノム、リファレンスのgtfファイルを指定してランする。複数のアセンブリを評価する時はスペースで区切る
(-c transcripts1.fasta<space>transcripts2.fasta)。
rnaQUAST.py -c transcripts1.fasta transcripts2.fasta -r reference.fasta --gtf gene_coordinates.gtf -t 2 -o output
- -c File(s) with transcripts in FASTA format separated by space.
- -o Directory to store all results. Default is rnaQUAST_results/results_<datetime>.
- -r Single file with reference genome containing all chromosomes/scaffolds in FASTA format (preferably with *.fasta, *.fa, *.fna, *.ffn or *.frn extension) OR *.txt file containing the one-per-line list of FASTA files with reference sequences.
- --gtf File with gene coordinates in GTF/GFF format (needs information about parent relations). We recommend to use files downloaded from GENCODE or Ensembl.
- -t Maximum number of threads. Default is min(number of CPUs / 2, 16).
- --blat Run with BLAT alignment tool instead of GMAP.
- --gene_mark Run with GeneMarkS-T gene prediction tool. Use --prokaryote option if the genome is prokaryotic.
レポートがPDF形式で出力される。

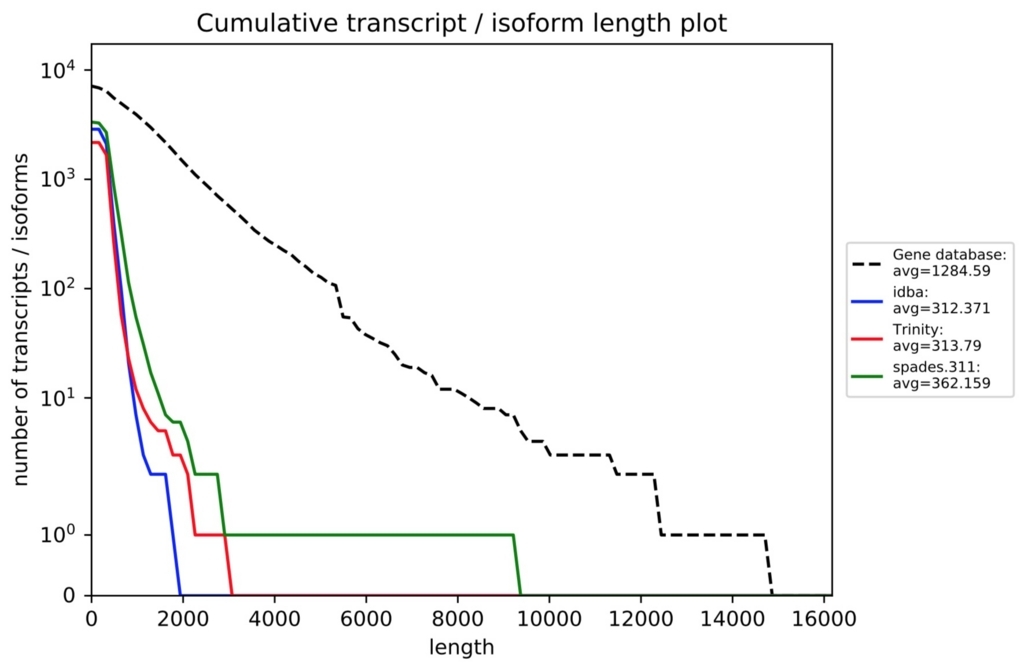
各評価項目の詳細はrnaQUAST 1.5 Manualから確認してください。オプション解説の下のほうに載ってます。
追記
buscoも走らせる場合、busco v3のHP(link)からダウンロードして解答したlineageディレクトリを指定する。
rnaQUAST.py -1 R1.fastq -2 R2.fastq -t 12 -o quast \
-c Trinity.fasta transcripts.fasta \
--busco_lineage protists_ensembl/
#reference genomeとアノテーション情報も利用できるなら使う
rnaQUAST.py -1 R1.fastq -2 R2.fastq -t 12 -o quast \
-c Trinity.fasta transcripts.fasta \
--busco_lineage protists_ensembl/
-r ref_genome.fa --gtf ref_annotation.gtf
引用
rnaQUAST: a quality assessment tool for de novo transcriptome assemblies.
Bushmanova E, Antipov D, Lapidus A, Suvorov V, PrjibelskiAD.
Bioinformatics. 2016 Jul 15;32(14):2210-2.
関連
ゲノムアセンブルの評価ツール QUAST
参考
Tools for building de novo transcriptome assembly
https://www.sciencedirect.com/science/article/pii/S2214662817301032