MViewは、シーケンスデータベース検索またはマルチアライメントの結果を抽出および再フォーマットし、オプションでWebページレイアウト用のHTMLマークアップを追加するコマンドラインユーティリティである。 一般的な形式に変換するためのフィルターとしても使用できる。
HP
Manual
https://desmid.github.io/mview/manual/manual.html
インストール
download
http://desmid.github.io/mview/index.html#download
1行目をperlのパスに修正する。
#!/usr/bin/perl
ここではmview-1.66のフォルダを/usr/local/に移動し、25行を以下のように修正した。
use lib '/usr/local/mview-1.66/lib';

> /usr/local/mview-1.66/bin/mview -h
$ /usr/local/mview-1.66/bin/mview -h
usage: mview [options] [file...]
Option names and parameter values can generally be abbreviated. Alternative
parameter values are listed in braces {}, followed by the default value in
square brackets [].
Some options take multiple arguments which must be supplied as a comma
separated list, like '1,8,9,10'. Subranges are allowed, so you could also
write that as '1,8:10' or even '1,8..10'. Any argument must be quoted if it
contains whitespace or a wildcard that might be expanded by the shell.
Option processing can be terminated using '--'.
Input/output formats:
-in format Input {blast,uvfasta,clustal,fasta,pir,msf,plain,hssp,maf,multas,mips,jnetz}.
-out format Output {pearson,fasta,pir,plain,clustal,msf,mview,rdb}. [mview]
Main formatting options:
-ruler on|off Show ruler. [on]
-alignment on|off Show alignment. [on]
-conservation on|off Show clustal conservation line. [off]
-consensus on|off Show consensus. [off]
-width columns Paginate alignment in blocks of width {N,full}. [full]
Percent identity calculations and filters:
-pcid mode Compute percent identities with respect to {aligned,reference,hit}. [aligned]
-reference string Use row N or row identifier as %identity reference. [query]
-minident N Only report sequences with percent identity >= N compared to reference. [0]
-maxident N Only report sequences with percent identity <= N compared to reference. [100]
-sort mode Resort output by coverage or percent identity {cov,pid,cov:pid,pid:cov,none}. [none]
General row/column filters:
-top count Report top N hits {N,all}. [all]
-show str[,str] Keep rows 1..N or identifiers.
-hide str[,str] Hide rows 1..N or identifiers.
-nops str[,str] Exclude rows 1..N or identifiers from calculations.
-range M:N,all Display column range M:N as numbered by ruler. [all]
Molecule type:
-moltype mol Affects coloring and format converions {aa,na,dna,rna}. [aa]
Alignment coloring:
-coloring mode Basic style of coloring {none,any,identity,mismatch,consensus,group}. [none]
-colormap name Name of colormap to use {see manual}. [P1]
-groupmap name Name of groupmap to use if coloring by consensus {see manual}. [P1]
-threshold N Threshold percentage for consensus coloring. [70]
-ignore mode Ignore singleton or class groups {none,class,singleton}. [none]
Consensus coloring:
-con_coloring mode Basic style of coloring {none,any,identity}. [none]
-con_colormap name Name of colormap to use {see manual}. [PC1]
-con_groupmap name Name of groupmap to use if coloring by consensus {see manual}. [P1]
-con_threshold N[,N] Consensus line thresholds. [100,90,80,70]
-con_ignore mode Ignore singleton or class groups {none,class,singleton}. [none]
-con_gaps on|off Count gaps during consensus computations if set to 'on'. [on]
Motif colouring:
-find pattern Find and highlight exact string or simple regular expression or ':' delimited set of patterns.
Miscellaneous formatting:
-label0 Switch off label {0= row number}. [set]
-label1 Switch off label {1= identifier}. [set]
-label2 Switch off label {2= description}. [set]
-label3 Switch off label {3= scores}. [set]
-label4 Switch off label {4= percent coverage}. [set]
-label5 Switch off label {5= percent identity}. [set]
-label6 Switch off label {6= first sequence positions: query}. [set]
-label7 Switch off label {7= second sequence positions: hit}. [set]
-label8 Switch off label {8= trailing fields}. [set]
-gap char Use this gap character. [-]
-sequences on|off Output sequences. [on]
-register on|off Output multi-pass alignments with columns in register. [on]
HTML markup:
-html mode Controls amount of HTML markup {head,body,data,full,off}. [off]
-bold Use bold emphasis for coloring sequence symbols. [unset]
-css mode Use Cascading Style Sheets {off,on,file:,http:}. [off]
-title string Page title string.
-pagecolor color Page backgound color. [white]
-textcolor color Page text color. [black]
-alncolor color Alignment background color. [white]
-labcolor color Alignment label color. [black]
-symcolor color Alignment symbol default color. [#666666]
-gapcolor color Alignment gap color. [#666666]
Database links:
-srs on|off Try to use sequence database links. [off]
-linkcolor color Link color. [blue]
-alinkcolor color Active link color. [red]
-vlinkcolor color Visited link color. [purple]
NCBI BLAST (series 1), WashU-BLAST:
-hsp mode HSP tiling mode {ranked,discrete,all}. [ranked]
-maxpval N,unlimited Ignore hits with p-value greater than N. [unlimited]
-minscore N,unlimited Ignore hits with score less than N. [unlimited]
-strand strands Report only these query strand orientations {p,m,both,*}. [both]
-keepinserts on|off Keep hit sequence insertions in unaligned output. [off]
NCBI BLAST (series 2), BLAST+:
-hsp mode HSP tiling mode {ranked,discrete,all}. [ranked]
-maxeval N,unlimited Ignore hits with e-value greater than N. [unlimited]
-minbits N,unlimited Ignore hits with bits less than N. [unlimited]
-strand strands Report only these query strand orientations {p,m,both,*}. [both]
-keepinserts on|off Keep hit sequence insertions in unaligned output. [off]
NCBI PSI-BLAST:
-hsp mode HSP tiling mode {ranked,discrete,all}. [ranked]
-maxeval N,unlimited Ignore hits with e-value greater than N. [unlimited]
-minbits N,unlimited Ignore hits with bits less than N. [unlimited]
-cycle cycles Process the N'th cycle of a multipass search {1..N,first,last,all,*}. [last]
-keepinserts on|off Keep hit sequence insertions in unaligned output. [off]
FASTA (U. of Virginia):
-minopt N,unlimited Ignore hits with opt score less than N. [unlimited]
-strand strands Report only these query strand orientations {p,m,both,*}. [both]
HSSP/Maxhom:
-chain chains Report only these chain names/numbers {A..B,1..N,first,last,all,*}. [all]
UCSC MAF:
-block blocks Report only these blocks {1..N,first,last,all,*}. [all]
MULTAL/MULTAS:
-block blocks Report only these blocks {1..N,first,last,all,*}. [all]
User defined colormap and consensus group definition:
-colorfile file Load more colormaps from file.
-groupfile file Load more groupmaps from file.
More information and help:
-help This help.
-listcolors Print listing of known colormaps.
-listgroups Print listing of known consensus groups.
-listcss Print style sheet.
MView 1.66, Copyright (C) 1997-2019 Nigel P. Brown
実行方法
ここでは、多様な機能のうち、multiple sequence alignmentの結果を受け取り、html形式で出力する手順を中心に記載する。
mview -html head -in fasta input_alignment_file > alignment.html
- -html <mode> Controls amount of HTML markup {head, body, data, full, off}. [off]
- -in <format> Input {blast, uvfasta, clustal, fasta, pir, msf, plain, hssp, maf, multas, mips,jnetz}.
- -out <format> Output {pearson,fasta,pir,plain,clustal,msf,mview,rdb}. [mview]
出力
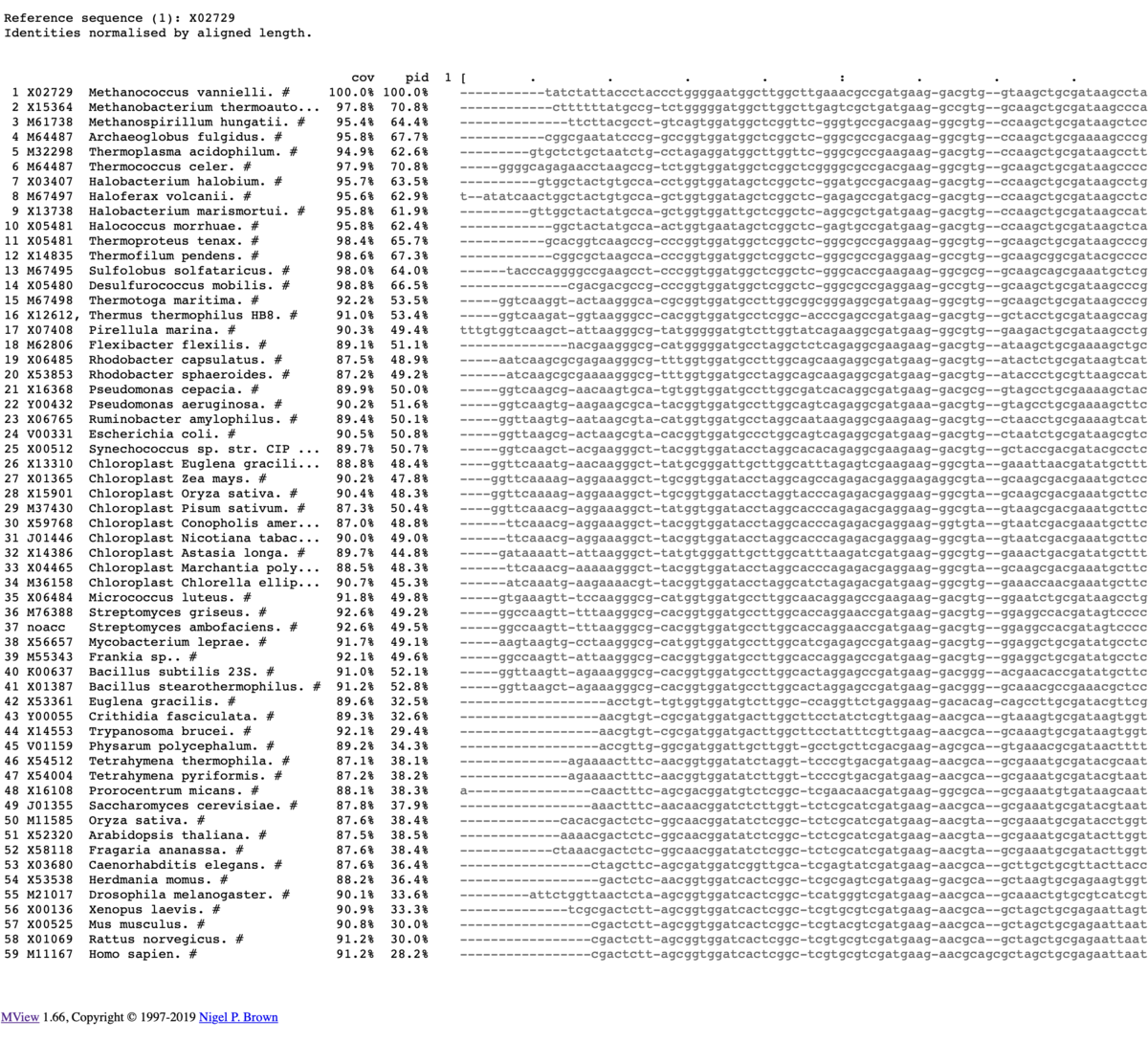
カラー出力
mview -html head -coloring any -bold -in fasta \
input_alignment_file > alignment.html
- -coloring <mode> Basic style of coloring {none, any, identity, mismatch, consensus, group}. [none]
- -bold Use bold emphasis for coloring sequence symbols. [unset]
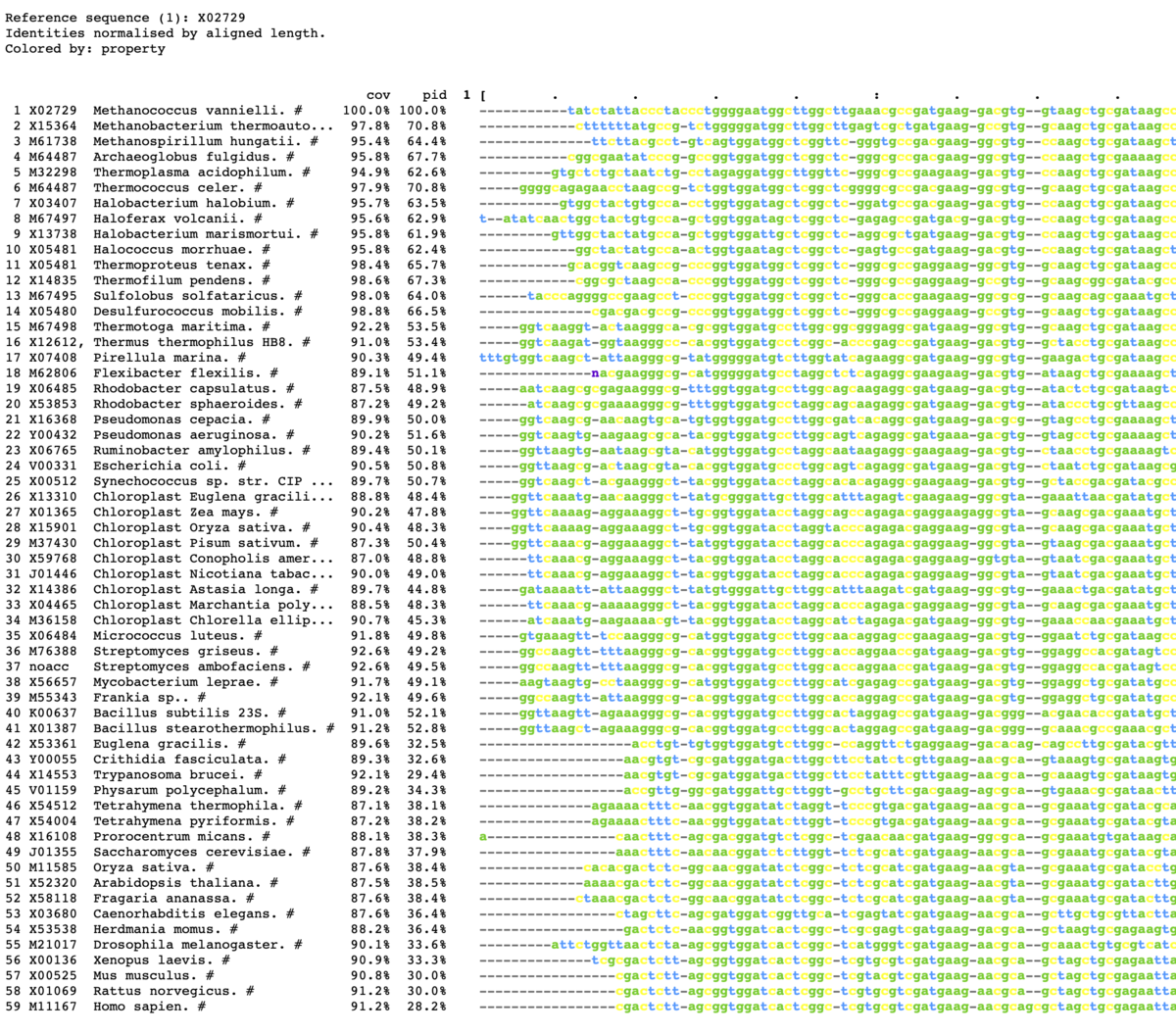
mview -html head -coloring any -bold -css on -in fasta \
input_alignment_file > alignment.html
- -css <mode> Use Cascading Style Sheets {off,on,file:,http:}. [off]
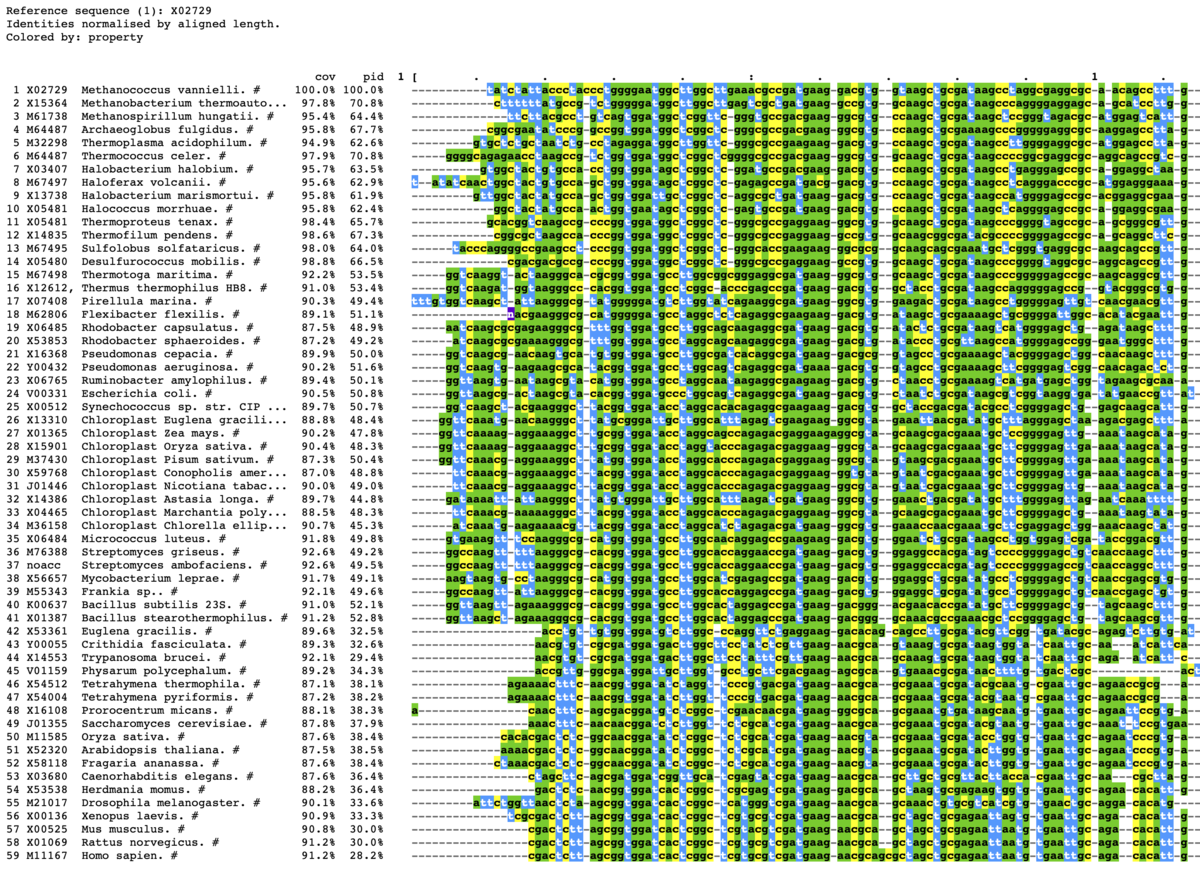
上から5つだけ出力する。範囲は1-100に限定する。
mview -html head -coloring identity -moltype dna \
-top 5 -range 1:100 -bold -css on \
-in fasta input_alignment_file > alignment.html
- -top <count> Report top N hits {N,all}. [all]
-
-range M:N,all Display column range M:N as numbered by ruler. [all]
-
-moltype <mol> Affects coloring and format converions {aa,na,dna,rna}. [aa]
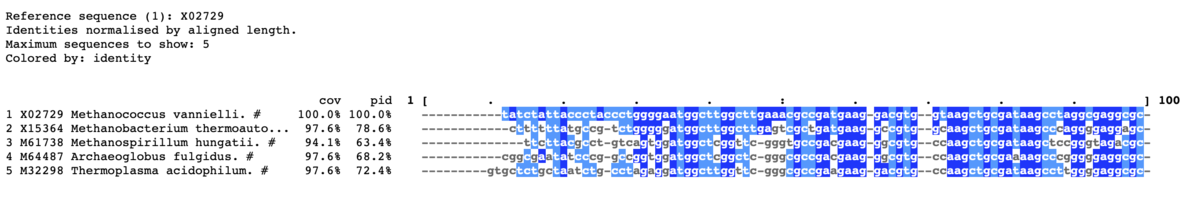
コンセンサス行を追加。パーセントも指定するなら"-con_threshold <NUM>"も追加する。
mview -html head -coloring identity -moltype dna \
-top 5 -range 1:100 -bold -css on -consensus on\
-in fasta input_alignment_file > alignment.html
- -consensus on|off Show consensus. [off]
- -con_threshold N[,N] Consensus line thresholds. [100,90,80,70]

8以上に色をつける。上から10表示。
mview -html head -coloring identity -moltype dna \
-top 10 -range 50:80 -bold -css on -ref 8\
-in fasta input_alignment_file > alignment.html
- -reference <string> Use row N or row identifier as %identity reference. [query]
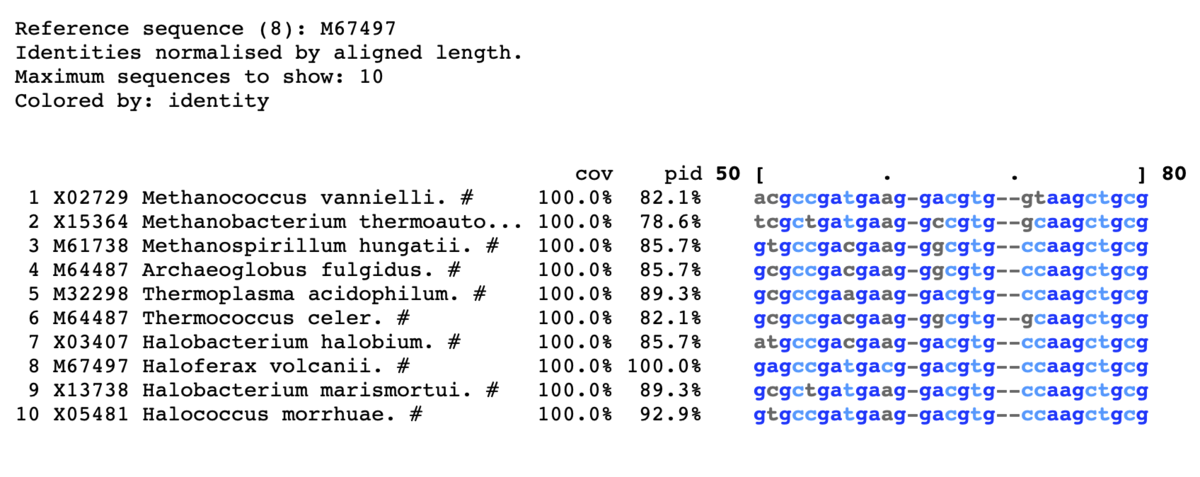
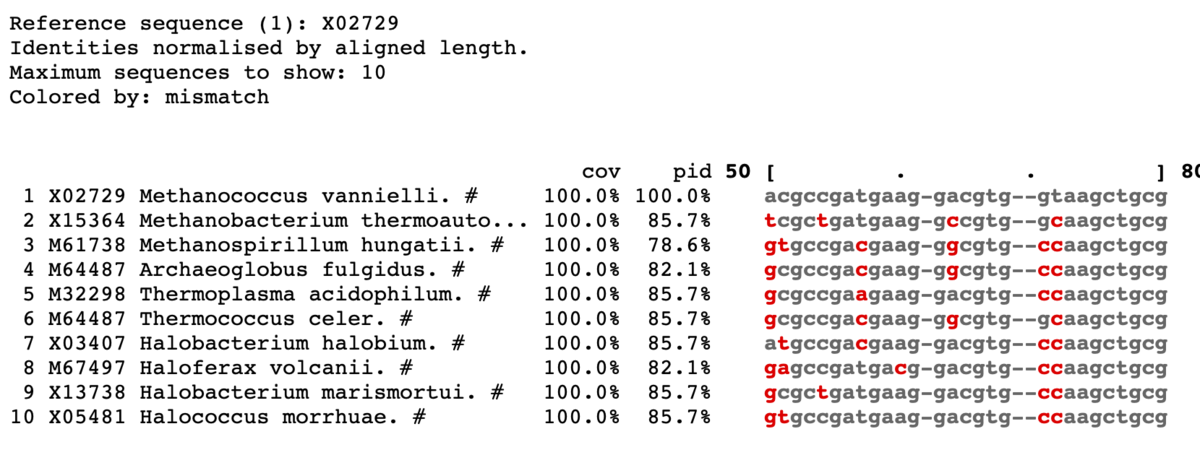
ミスマッチを赤で表示。
mview -html head -coloring identity -moltype dna -top 10 \
-range 50:80 -bold -css on -coloring mismatch -colormap red \
-in fasta input_alignment_file > alignment.html
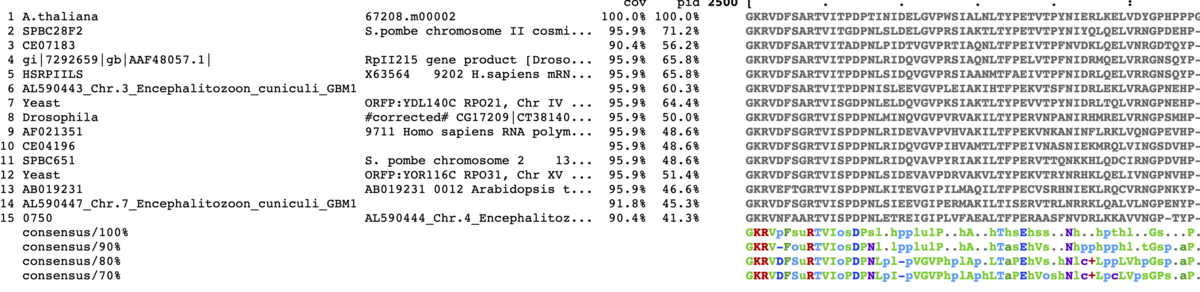
TOP15表示。コンセンサス配列を一番下に表示。90%以上。入力はprotein配列に変更。
mview -html head -coloring identity -moltype aa -top 15 \
-range 2500:2580 -bold \
-threshold 90 -consensus on -con_threshold 90 \
-in fasta input_alignment_file > alignment.html
- -threshold <N> Threshold percentage for consensus coloring. [70]
- -consensus on|off Show consensus. [off]
- -con_threshold N[,N] Consensus line thresholds. [100,90,80,70]

特定のアミノ酸配列だけカラー表示。GVP。
mview -html head -coloring identity -moltype aa -top 15 \
-range 2500:2580 -bold \
-find GVP \
-in fasta input_alignment_file > alignment.html
- -find <pattern> Find and highlight exact string or simple regular expression or ':' delimited set of patterns.

正規表現も認識する。
コンセンサス配列をカラー表示。
mview -html head -coloring identity -moltype aa -top 15 \
-range 2500:2580 -bold \
-consensus on -con_coloring any \
-in fasta input_alignment_file > alignment.html
- -con_coloring <mode> Basic style of coloring {none,any,identity}. [none]
カラーキーをclustalに変更。
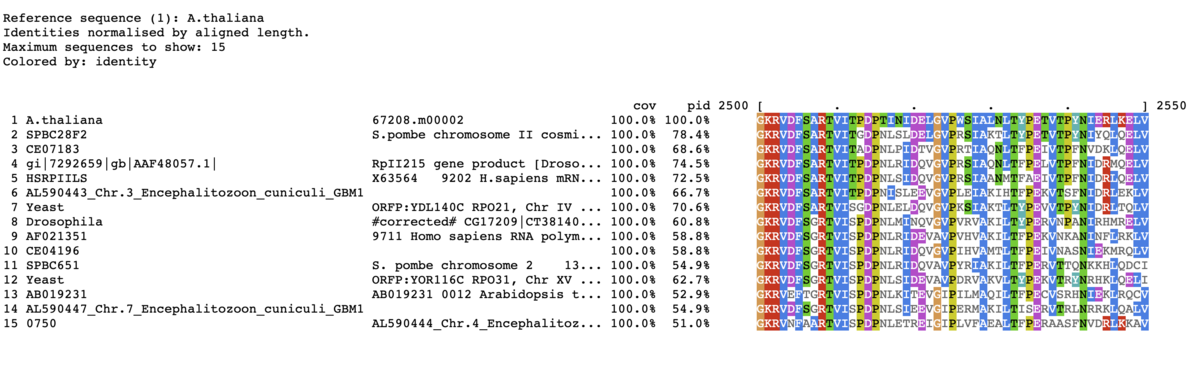
mview -html head -coloring id -colormap -moltype aa \
-css on clustal -top 15 -range 2500:2550 \
-in fasta input_alignment_file > alignment.html
引用
MView: a web-compatible database search or multiple alignment viewer
Brown NP, Leroy C, Sander C
Bioinformatics. 1998;14(4):380-1
関連