RNA-Seqは特定のバイアス、アーティファクトを受けやすく、 堅牢で包括的なクオリティチェックが重要になる。とくにサンプル調製、ライブラリー作成、またはシークエンシングのエラーは、 予期せぬアーティファクト、バイアスを引き起こす。適切に処理できるように、そのような問題を検出することが重要になるが、品質を自動的にテストする包括的な方法が存在しない。
QoRTsは幅広いクオリティマトリクスを生成し、インフォマティシャンが適切にクオリティチェックを行えるようサポートする。
公式サイト
QoRTs: Quality of RNA-Seq Toolset
Example Walkthrough
QoRTs/example-walkthrough.pdf at master · hartleys/QoRTs · GitHub
インストール
Githubのリリースからコンパイル済みのjarファイルをダウンロードできる。マニュアルもある。
https://github.com/hartleys/QoRTs/releases
java -jar QoRTs.jar QC -man #マニュアル
#描画するためのRのパッケージも入れておく。Rに入って
install.packages("http://hartleys.github.io/QoRTs/QoRTs_LATEST.tar.gz", repos=NULL, type="source");
r$ java -jar QoRTs.jar QC --man
Starting QoRTs v1.3.0 (Compiled Fri Oct 20 11:56:37 EDT 2017)
Starting time: (Mon Jan 22 17:49:51 JST 2018)
NAME
QC
Version: 1.3.0 (Updated Fri Oct 20 11:56:37 EDT 2017)
USAGE
java [Java Options] -jar QoRTs.jar QC [options] infile
gtffile.gtf qcDataDir
DESCRIPTION:
This utility runs a large battery of QC / data processing tools
on a single given sam or bam file. This is the primary function
of the QoRTs utility. All analyses are run via a single pass
through the sam/bam file.
REQUIRED ARGUMENTS:
infile
The input .bam or .sam file of aligned sequencing reads. Or
'-' to read from stdin.
(String)
gtffile.gtf
The gtf annotation file. This tool was designed to use the
standard gtf annotations provided by Ensembl, but other
annotations can be used as well.
If the filename ends with ".gz" or ".zip", the file will be
parsed using the appropriate decompression method.
(String)
qcDataDir
The output directory.
(String)
OPTIONS:
--singleEnded
Flag to indicate that reads are single end.
(flag)
--stranded
Flag to indicate that data is stranded.
(flag)
--stranded_fr_secondstrand
Flag to indicate that reads are from a fr_secondstrand type
of stranded library (equivalent to the "stranded = yes"
option in HTSeq or the "fr_secondStrand" library-type option
in TopHat/CuffLinks). If your data is stranded, you must
know the library type in order to analyze it properly. This
utility uses the same definitions as cufflinks to define
strandedness type. By default, the fr_firststrand library
type is assumed for all stranded data (equivalent to the
"stranded = reverse" option in HTSeq).
(flag)
--maxReadLength len
Sets the maximum read length. For unclipped datasets this
option is not necessary since the read length can be
determined from the data. By default, QoRTs will attempt to
determine the max read length by examining the first 1000
reads. If your data is hard-clipped prior to alignment, then
it is strongly recommended that this option be included, or
else an error may occur. Note that hard-clipping data prior
to alignment is generally not recommended, because this
makes it difficult (or impossible) to determine the
sequencer read-cycle of each nucleotide base. This may
obfuscate cycle-specific artifacts, trends, or errors, the
detection of which is one of the primary purposes of QoRTs!
In addition, hard clipping (whether before or after
alignment) removes quality score data, and thus quality
score metrics may be misleadingly optimistic. A MUCH
preferable method of removing undesired sequence is to
replace such sequence with N's, which preserves the quality
score and the sequencer cycle information while still
removing undesired sequence.
(Int)
--minMAPQ num
Filter out reads with less than the given MAPQ. Most RNA-Seq
aligners use the MAPQ field to differentiate uniquely-mapped
and multi-mapped reads. However, different aligners use a
different MAPQ conventions. By default, all reads with a
MAPQ of less than 255 will be excluded, as this is the MAPQ
associated with uniquely-aligned reads generated by the
RNA-STAR aligner. For use with TopHat2 you should set this
to 50. The MAPQ behavior for GSNAP is not well documented,
but it appears that a filtering threshold of 30 should be
adequate. Set this to 0 to turn off mapq filtering.
(Int)
--generatePlots
Generate all single-replicate QC plots. Equivalent to the
combination of: --generateMultiPlot --generateSeparatePlots
and --generatePdfReport. This option will cause QoRTs to
make an Rscript system call, loading the R package QoRTs.
(Note: this requires that R be installed and in the PATH,
and that QoRTs be installed on that R installation)
(flag)
--testRun
Flag to indicate that only the first 100k reads should be
read in. Used for testing.
(flag)
--keepMultiMapped
Flag to indicate that the tool should NOT filter out
multi-mapped reads. Note that even with this flag raised
this utility will still only use the 'primary' alignment
location for each read. By default any reads that are marked
as multi-mapped will be ignored entirely. Most aligners use
the MAPQ value to mark multi-mapped reads. Any read with
MAPQ < 255 is assumed to be non-uniquely mapped (this is the
standard used by RNA-STAR and TopHat/TopHat2). This option
is equivalent to "--minMAPQ 0".
(flag)
--noGzipOutput
Flag to indicate that output files should NOT be compressed
into the gzip format. By default almost all output files are
compressed to save space.
(flag)
--readGroup readGroupName
If this option is set, all analyses will be restricted to
ONLY reads that are tagged with the given readGroupName
(using an RG tag). This can be used if multiple read-groups
have already been combined into a single bam file, but you
want to summarize each read-group separately.
(String)
--dropChrom dropChromosomes
A comma-delimited list of chromosomes to ignore and exclude
from all analyses. Important: no whitespace!
(CommaDelimitedListOfStrings)
--skipFunctions func1,func2,...
A list of functions to skip (comma-delimited, no
whitespace). See the sub-functions list, below. The
default-on functions are: NVC, GCDistribution, GeneCalcs,
readLengthDistro, QualityScoreDistribution,
writeJunctionSeqCounts, writeKnownSplices,
writeNovelSplices, writeClippedNVC, CigarOpDistribution,
overlapMatch, cigarLocusCounts, InsertSize, chromCounts,
writeSpliceExon, writeGenewiseGeneBody, JunctionCalcs,
writeGeneCounts, writeBiotypeCounts, writeDESeq,
writeDEXSeq, writeGeneBody, StrandCheck
(CommaDelimitedListOfStrings)
--addFunctions func1,func2,...
A list of functions to add (comma-delimited, no whitespace).
This can be used to add functions that are off by default.
Followed by a comma delimited list, with no internal
whitespace. See the sub-functions list, below. The
default-off functions are: mismatchEngine,
annotatedSpliceExonCounts, calcOnTarget, FPKM, cigarMatch,
testDataDump, writeGeneBodyIv, fastqUtils, referenceMatch,
writeDocs, makeJunctionBed, makeWiggles,
makeAllBrowserTracks, calcDetailedGeneCounts
(CommaDelimitedListOfStrings)
--runFunctions func1,func2,...
The complete list of functions to run (comma-delimited, no
whitespace). Setting this option runs ONLY for the functions
explicitly requested here (along with any functions upon
which the assigned functions are dependent). See the
sub-functions list, below. Allowed options are: NVC,
mismatchEngine, annotatedSpliceExonCounts, GCDistribution,
calcOnTarget, GeneCalcs, FPKM, readLengthDistro, cigarMatch,
QualityScoreDistribution, testDataDump,
writeJunctionSeqCounts, writeKnownSplices,
writeNovelSplices, writeClippedNVC, CigarOpDistribution,
overlapMatch, cigarLocusCounts, InsertSize, chromCounts,
writeGeneBodyIv, fastqUtils, writeSpliceExon,
referenceMatch, writeGenewiseGeneBody, JunctionCalcs,
writeGeneCounts, writeDocs, makeJunctionBed,
writeBiotypeCounts, writeDESeq, writeDEXSeq, makeWiggles,
writeGeneBody, StrandCheck, makeAllBrowserTracks,
calcDetailedGeneCounts
(CommaDelimitedListOfStrings)
--seqReadCt val
(Optional) The total number of reads (or read-pairs, for
paired-end data) generated by the sequencer for this sample,
prior to alignment. This will be passed on into the
QC.summary.txt file and used to calculate mapping rate.
(Int)
--rawfastq myfastq.1.fq.gz,myfastq.2.fq.gz
(Optional) The raw fastq, prior to alignment. In normal
operation, this is used ONLY to calculate the number of
pre-alignment reads (or read-pairs) simply by counting the
number of lines and dividing by 4. Alternatively, the number
of pre-alignment read-pairs can be included explicitly via
the --seqReadCt option, or added in the plotting /
cross-comparison step by including the input.read.pair.count
column in the replicate decoder.In general, the --seqReadCt
option is recommended when available.
Certain optional QC functions are also available that
utilize the raw Fastq file in other ways. If the filename
ends with ".gz" or ".zip", the file will be parsed using the
appropriate decompression method.
(CommaDelimitedListOfStrings)
--chromSizes chrom.sizes.txt
A chrom.sizes file. The first (tab-delimited) column must
contain all chromosomes found in the dataset. The second
column must contain chromosome sizes (in base-pairs). If a
standard genome is being used, it is strongly recommended
that this be generated by the UCSC utility
'fetchChromSizes'.
This file is ONLY needed to produce wiggle files. If this is
provided, then by default QoRTs will produce 100-bp-window
wiggle files (and junction '.bed' files) for the supplied
data.In order to produce wiggle files, this parameter is
REQUIRED.
(String)
--title myTitle
The title of the replicate. Used for the track name in the
track definition line of any browser tracks ('.wig' or
'.bed' files) generated by this utility. Also may be used in
the figure text, if figures are being generated.Note that no
browser tracks will be created by default, unless the
'--chromSizes' option is set. Bed files can also be
generated using the option '--addFunction makeJunctionBed'
(String)
--flatgff flattenedGffFile.gff.gz
A "flattened" gff file that matches the standard gtf file.
Optional. The "flattened" gff file assigns unique
identifiers for all exons, splice junctions, and
aggregate-genes. This is used for the junction counts and
exon counts (for DEXSeq). The flattened gtf file can be
generated using the "makeFlatGff" command. Flattened GFF
files containing novel splice junctions can be generated
using the "mergeNovelSplices" function. Note that (for most
purposes) the command should be run with the same
strandedness code as found in the dataset. Running a
flattened gff that was generated using a different
strandedness mode may be useful for certain purposes, but is
generally not supported and is for advanced users only.See
the documentation for makeFlatGff for more information.
If the filename ends with ".gz" or ".zip", the file will be
parsed using the appropriate decompression method.
(String)
--generateMultiPlot
Generate a multi-frame figure, containing a visual summary
of all QC stats. (Note: this requires that R be installed
and in the PATH, and that QoRTs be installed on that R
installation)
(flag)
--generateSeparatePlots
Generate seperate plots for each QC stat, rather than only
one big multiplot. (Note: this requires that R be installed
and in the PATH, and that QoRTs be installed on that R
installation)
(flag)
--generatePdfReport
Generate a pdf report. (Note: this requires that R be
installed and in the PATH, and that QoRTs be installed on
that R installation)
(flag)
--adjustPhredScore val
QoRTs expects input files to conform to the SAM format
specification, which requires all Phred scores to be in
Phred+33 encoding. However some older tools produce SAM
files with nonstandard encodings. To read such data, you can
set this parameter to subtract from the apparent (phred+33)
phred score. Thus, to read Phred+64 data (produced by
Illumina v1.3-1.7), set this parameter to 31. Note: QoRTs
does not support negative Phred scores. NOTE: THIS OPTION IS
EXPERIMENTAL!
(Int)
--maxPhredScore val
According to the standard FASTQ and SAM format
specification, Phred quality scores are supposed to range
from 0 to 41. However, certain sequencing machines such as
the HiSeq4000 supposedly produce occasional quality scores
as high as 45. If your dataset contains quality scores in
excess of 41, then you must use this option to set the
maximum legal quality score. Otherwise, QoRTs will throw an
error.
(Int)
--summaryFileSuffix .summary.txt
The suffix of the 'summary' file. This is useful to set if
you want to run multiple QC runs in parallel to reduce
runtime, without overwriting one another's summary files.In
particular, the NVC metrics often take a long time to run,
so splitting those off using the --runFunctions parameter
might speed things up considerably. Note that 'QC' will be
appended in the actual filename. THIS OPTION IS BETA!
(String)
--extractReadsByMetric metric=value
THIS OPTIONAL PARAMETER IS STILL UNDER BETA TESTING. This
parameter allows the user to extract anomalous reads that
showed up in previous QoRTs runs. Currently reads can be
extracted based on the following metrics: StrandTestStatus,
InsertSize and GCcount.
(String)
--keepOnlyOnTarget
Experimental flag. Ignores reads that DO NOT fall within the
target region (specified by the required bedfile using the
--targetRegionBed parameter).
(flag)
--dropOnTarget
Experimental flag. Ignores reads that DO fall within the
target region (specified by the required bedfile using the
--targetRegionBed parameter).
(flag)
--randomSubsample 1.00
If this option is set, QoRTs will ignore a random fraction
of the input read pairs. This can drastically reduce
runtime, though it may reduce the accuracy of the output QC
metrics.
(Double)
--restrictToGeneList geneList.txt
If this option is set, almost all analyses will be
restricted to reads that are found on genes named in the
supplied gene list file. The file should contain a gene ID
on each line and nothing else. The only functions that will
be run on the full set of all reads will be the functions
that calculate the gene mapping itself. NOTE: if you want to
include ambiguous reads, include a line with the text:
'_ambiguous'. If you want to include reads that do not map
to any known feature, include a line with the text:
'_no_feature'. WARNING: this is not intended for default
use. It is intended to be used when re-running QoRTs, with
the intention of examining artifacts that can be caused in
various plots by a small number of genes with extremely high
coverage. For example, GC content plots sometimes contain
visible spikes caused by small mitochondrial genes with
extremely high expression.ADDITIONAL WARNING: This feature
is in BETA, and is not yet fully tested.
(String)
--dropGeneList geneList.txt
If this option is set, almost all analyses will be
restricted to reads that are NOT found on genes named in the
supplied gene list file. The file should contain a gene ID
on each line and nothing else. The only functions that will
be run on the full set of all reads will be the functions
that calculate the gene mapping itself. NOTE: if you want to
EXCLUDE ambiguous reads, include a line with the text:
'_ambiguous'. If you want to EXCLUDE reads that do not map
to any known feature, include a line with the text:
'_no_feature'. WARNING: this is not intended for default
use. It is intended to be used when re-running QoRTs, with
the intention of examining artifacts that can be caused by
certain individual 'problem genes'. For example, GC content
plots sometimes contain visible spikes caused by small
transcripts / RNA's with extremely high expression
levels.ADDITIONAL WARNING: This feature is in BETA, and is
not yet fully tested.
(String)
--DNA
BETA: This flag makes various changes to allow QoRTs to run
on whole-exome or whole-genome DNA-Seq data.
(flag)
--RNA
Indicates that the data is RNA-Seq (this is the default:
flag does nothing).
(flag)
--genomeFA chr.fa.gz[,chr2.fa,...]
Reference genome sequence. This can either be a single FASTA
file with all the chromosomes included, or a comma-delimited
list of fasta files with 1 chromosome each. Note: IF
multiple fasta files are specificed, each must contain ONLY
ONE chromosome. If a single multi-chromosome fasta file is
specificed, performance will be improved if the chromosomes
are in the same order as they are found in the BAM file,
however, this is not required. The genomic sequence is used
by certain experimental sub-utilities (currently only the
referenceMatch utility). Comma delimited, no spaces. Fasta
files can be in plaintext, gzipped or zipped.
(CommaDelimitedListOfStrings)
--genomeBufferSize val
The size of the genome fasta buffer. Tuning this parameter
may improve performance.
(Int)
--outfilePrefix sampID
Prefix to be prepended to all output files. If this is set,
all output files will use the format:
"outfiledir/<prefix>QC.qcfilename.txt.gz"
(String)
--nameSorted
DEPRECATED: Relevant for paired-end reads only.
This flag is used to run QoRTs in "name-sorted" mode. This
flag is optional, as under the default mode QoRTs will
accept BAM files sorted by either name OR position. However,
The only actual requirement in this mode is that read pairs
be adjacent.
Errors may occur if the SAM flags are inconsistent: for
example, if orphaned reads appear with the "mate mapped" SAM
flag set.
(flag)
--coordSorted
DEPRECATED: this mode is now subsumed by the default mode
and as such this parameter is now nonfunctional.
Note that, in the default mode, for paired-end data QoRTs
will accept EITHER coordinate-sorted OR name-sorted bam
files. In "--nameSorted" mode, QoRTs ONLY accepts
name-sorted bam files.
If a large fraction of the read-pairs are mapped to
extremely distant loci (or to different chromosomes), then
memory issues may arise. However, this should not be a
problem with most datasets. Technically by default QoRTs can
run on arbitrarily-ordered bam files, but this is STRONGLY
not recommended, as memory usage will by greatly increased.
(flag)
--fileContainsNoMultiMappedReads
DEPRECATED. Flag to indicate that the input sam/bam file
contains only primary alignments (ie, no multi-mapped
reads). This flag is ALWAYS OPTIONAL, but when applicable
this utility will run (slightly) faster when using this
argument. (DEPRECIATED! The performance improvement was
marginal)
(flag)
--parallelFileRead
DEPRECATED: DO NOT USE. Flag to indicate that bam file
reading should be run in paralell for increased speed. Note
that in this mode you CANNOT read from stdin. Also note that
for this to do anything useful, the numThreads option must
be set to some number greater than 1. Also note that
additional threads above 9 will have no appreciable affect
on speed.
(flag)
--numThreads num
DEPRECIATED, nonfunctional.
(Int)
--checkForAlignmentBlocks
Certain aligners will mark reads 'aligned' even though they
have no aligned bases. This option will automatically check
for some reads and ignore them, rather than throwing an
error.
(flag)
--targetRegionBed targetRegion.bed
For whole exome sequencing, this specifies the exome target
regions.
(String)
--stopAfterNReads n
Stop after reading in n reads or read-pairs.
(Int)
--randomSeed n
Use specified random seed.
(Long)
--parseIlluminaStyleReadIDs
Specifies that the read-names are in the illumina style.
CURRENTLY NONFUNCTIONAL!
(flag)
--verbose
Flag to indicate that debugging information and extra
progress information should be sent to stderr.
(flag)
--quiet
Flag to indicate that only errors and warnings should be
sent to stderr.
(flag)
DEFAULT SUB-FUNCTIONS
NVC
Nucleotide-vs-Cycle counts.
GCDistribution
Calculate GC content distribution.
GeneCalcs
Find gene assignment and gene body calculations.
readLengthDistro
Tabulates the distribution of read lengths.
QualityScoreDistribution
Calculate quality scores by cycle.
writeJunctionSeqCounts
Write counts file designed for use with JunctionSeq
(contains known splice junctions, gene counts, and exon
counts). [Depends: writeSpliceExon]
writeKnownSplices
Write known splice junction counts. [Depends: JunctionCalcs]
writeNovelSplices
Write novel splice junction counts. [Depends: JunctionCalcs]
writeClippedNVC
Write NVC file containing clipped sequences. [Depends: NVC]
CigarOpDistribution
Cigar operation rates by cycle and cigar operation length
rates (deletions, insertions, splicing, clipping, etc).
overlapMatch
BETA: This function calculates the matching of overlapping
sections of paired reads. [Depends: mismatchEngine]
cigarLocusCounts
BETA: This function is still undergoing basic testing. It is
not intended for production use at this time.
InsertSize
Insert size distribution (paired-end data only).
chromCounts
Calculate chromosome counts
writeSpliceExon
Synonym for function "writeJunctionSeqCounts" (for
backwards-compatibility) [Depends: JunctionCalcs]
writeGenewiseGeneBody
Write file containing gene-body distributions for each
(non-overlapping) gene. [Depends: writeGeneBody]
JunctionCalcs
Find splice junctions (both novel and annotated).
writeGeneCounts
Write extended gene-level read/read-pair counts file
(includes counts for CDS/UTR, ambiguous regions, etc).
[Depends: GeneCalcs]
writeBiotypeCounts
Write a table listing read counts for each gene BioType
(uses the optional "gene_biotype" GTF attribute). [Depends:
GeneCalcs]
writeDESeq
Write gene-level read/read-pair counts file, suitable for
use with DESeq, EdgeR, etc. [Depends: GeneCalcs]
writeDEXSeq
Write exon-level read/read-pair counts file, designed for
use with DEXSeq. [Depends: JunctionCalcs]
writeGeneBody
Write gene-body distribution file. [Depends: GeneCalcs]
StrandCheck
Check the strandedness of the data. Note that if the
stranded option is set incorrectly, this tool will
automatically print a warning to that effect.
NON-DEFAULT SUB-FUNCTIONS
mismatchEngine
Internal module that runs overlap/reference mismatch
calculations. Automatically included on any runs that
include these functions.
annotatedSpliceExonCounts
Write counts for exons, known-splice-junctions, and genes,
with annotation columns indicating chromosome, etc (default:
OFF) [Depends: JunctionCalcs]
calcOnTarget
BETA: requires --targetRegionBed parameter. This function
calculates the rates at which reads intersect with the
On-Target area. Intended for whole exome sequencing data.
Make sure to use the --targetRegionBed parameter or else
this function will deactivate! (Default: ON iff
targetRegionBed param is found)
FPKM
Write FPKM values. Note: FPKMs are generally NOT the
recommended normalization method. We recommend using a more
advanced normalization as provided by DESeq, edgeR,
CuffLinks, or similar (default: OFF)
cigarMatch
Work-In-Progress: this function is a placeholder for future
functionality, and is not intended for use at this time.
(default: OFF)
testDataDump
EXPERIMENTAL: This function dumps a bunch of information for
internal testing purposes. NOT FOR GENERAL USE! (default:
OFF)
writeGeneBodyIv
Writes an optional additional file detailing the intervals
used in the gene-body coverage calculations
("QC.geneBodyCoverage.DEBUG.intervals.txt.gz") (default:
OFF) [Depends: writeGeneBody]
fastqUtils
BETA: requires --rawfastq parameter. Adds additional tests
that use the supplied raw fastq file. Requires that one (or
two) fastq files be supplied. (Default: ON iff rawfastq
param is found)
referenceMatch
BETA: requires --genomeFA parameter. This function
calculates the matching against the reference. Requires the
specification of genome fasta file(s). REQUIRES
COORDINATE-SORTED BAM FILES! REQUIRES THAT FA AND BAM HAVE
THE SAME CHROMOSOME ORDERING! (Default: ON iff genomeFA
param is found) [Depends: mismatchEngine]
writeDocs
Writes a QC.documentation.txt file that documents all output
files.
makeJunctionBed
Write splice-junction count "bed" files. (default: OFF)
makeWiggles
Write "wiggle" coverage files with 100-bp window size. Note:
this REQUIRES that the --chromSizes parameter be included!
(default: OFF)
makeAllBrowserTracks
Write both the "wiggle" and the splice-junction bed files
(default: OFF) [Depends: makeJunctionBed, makeWiggles]
calcDetailedGeneCounts
Calculate more detailed read counts for each gene, counting
the number of reads that cover introns, cross-strand, etc
(default: OFF)
AUTHORS:
Stephen W. Hartley, Ph.D. stephen.hartley (at nih dot gov)
LEGAL:
This software is "United States Government Work" under the terms
of the United States Copyright Act. It was written as part of
the authors' official duties for the United States Government
and thus cannot be copyrighted. This software is freely
available to the public for use without a copyright notice.
Restrictions cannot be placed on its present or future use.
Although all reasonable efforts have been taken to ensure the
accuracy and reliability of the software and data, the National
Human Genome Research Institute (NHGRI) and the U.S. Government
does not and cannot warrant the performance or results that may
be obtained by using this software or data. NHGRI and the U.S.
Government disclaims all warranties as to performance,
merchantability or fitness for any particular purpose.
In any work or product derived from this material, proper
attribution of the authors as the source of the software or data
should be made, using "NHGRI Genome Technology Branch" as the
citation.
NOTE: This package includes (internally) the sam-1.113.jar
library from picard tools. That package uses the MIT license,
which can be accessed using the command:
java -jar thisjarfile.jar help samjdkinfo
Done. (Mon Jan 22 17:49:51 JST 2018)
例えばbin/に入れて省略名"QoRTs"でランできるようにしておく。
mv QoRTs.jar /usr/local/bin/
echo alias QoRTs=\'java -jar /usr/local/bin/QoRTs.jar\' >> ~/.bash_profile && source ~/.bash_profile
ラン
アライメント結果のbamとgtfを指定してランする。結果は指定したディレクトリに出力される。
QoRTs --generatePdfReport input.bam input.gtf qcDataDir
- --singleEnded Flag to indicate that reads are single end. (flag)
- --stranded Flag to indicate that data is stranded. (flag)
- --stranded_fr_secondstrand Flag to indicate that reads are from a fr_secondstrand type of stranded library (equivalent to the "stranded = yes" option in HTSeq or the "fr_secondStrand" library-type option in TopHat/CuffLinks). If your data is stranded, you must know the library type in order to analyze it properly. This utility uses the same definitions as cufflinks to define strandedness type. By default, the fr_firststrand library type is assumed for all stranded data (equivalent to the "stranded = reverse" option in HTSeq). (flag)
- --generateMultiPlot Generate a multi-frame figure, containing a visual summary of all QC stats. (Note: this requires that R be installed and in the PATH, and that QoRTs be installed on that R installation) (flag)
- --generatePdfReport Generate a pdf report. (Note: this requires that R be installed and in the PATH, and that QoRTs be installed on that R installation)
シングルエンドのbamなら"--singleEnded"のフラグをつけてランする。結果は指定したディレクトリqcDataDir/に出力される。
--generatePdfReportをつけると、次のようなPDFレポートが出力される。
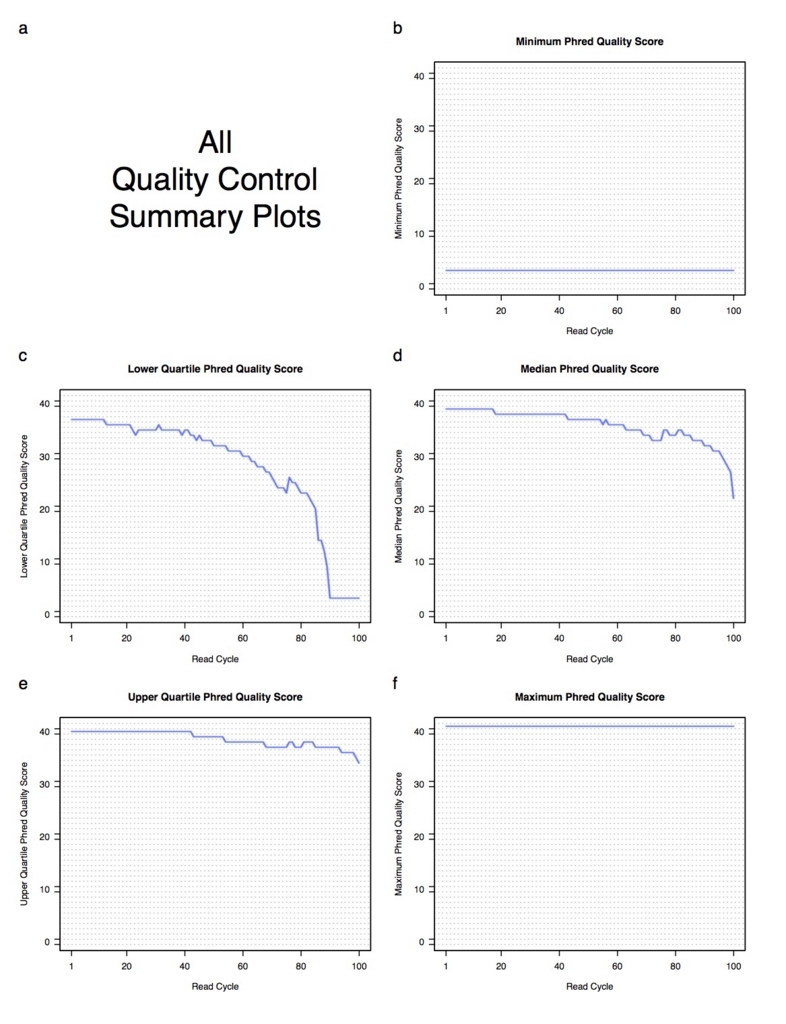





QCコマンド以外に、QC出力のreplicates内のマージ、UCSCで使えるカバレッジのwigファイルへの変換、レポートを出力するユーティリティコマンドなどいくつかあります。詳細は次のコマンドで確認してください。
QoRTs utilname --man
こちらにも分かりやすく説明されています。
QoRTs: Quality of RNA-Seq Toolset
引用
QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments
Stephen W. HartleyEmail author and James C. Mullikin
BMC Bioinformatics 201516:224







