2019 5/15 リンク、condaインストール追加
2019 5/16 タイトル修正
2020 9/27 help更新
ハイスループットの全ゲノムショットガン(WGS)データセットの迅速な解析は、大きなサイズが生み出す複雑さのためにチャレンジングである(Schatz et al、2012)。 WGSデータを分析するためのリファレンスが不要なアプローチは、基本的な品質、リード長、GCコンテンツ(Yang et al、2013)の調査とk-mer(サイズk のワード)スペクトラム探索を含む(Chor et al、2009; Lo and Chain、2014)。頻繁に使用されるリファレンスフリーの品質管理ツールはFastQC(http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)である。
K-merスペクトラムは、データ品質(エラーのレベル、シーケンシングバイアス、シーケンシングカバレッジおよび潜在的汚染)だけでなく、ゲノムの複雑さ(サイズ、核型、ヘテロ接合性および反復含有量; Simpson、2014)に関する情報を明らかにする。 WGSデータセットのペアワイズ比較(Anvar et al、2014)により、スペクトラムの違いを強調して問題のあるサンプルを識別できる追加情報を抽出できる。
K-mer analysis Tookitは、シーケンスデータから直接的に任意の長さのk-merのスペクトルを迅速にカウント、比較、分析するためのツール群である。任意の分布のk-merをフィルタリングする機能も備えている。
KAT's Documentation
Welcome to KAT’s documentation! — kat 2.4.1 documentation

マニュアルより転載。k-merスペクトラムを描くことで、ゲノムに関する様々な情報を得ることができる。図にはないが、たいていのシーケンスデータでは左端に最も大きいピークがある(おそらくシーケンスエラー由来)。
kat-comp introduction
k-mer counting, part I: Introduction
インストール
依存
- GCC V4.8+
- make
- autoconf V2.53+
- automake V1.11+
- libtool V2.4.2+
- pthreads (probably already installed)
- zlib
- Python V3.5+ with the tabulate, scipy, numpy and matplotlib packages and C API installed. Python is optional but highly recommended, without python, KAT functionality is limited: no plots, no distribution analysis, and no documentation.
- Sphinx-doc V1.3+ (Optional: only required for building the documentation.
ビルドはやや複雑だが、KATはbrewによるインストールがサポートされており、ワンライナーで導入できる。
brew install brewsci/bio/kat
#bioconda
conda install -c bioconda -y kat
$ kat
The K-mer Analysis Toolkit (KAT) contains a number of tools that analyse jellyfish K-mer hashes.
The First argument should be the tool/mode you wish to use:
* hist: Create an histogram of k-mer occurrences from a sequence file. Similar to
jellyfish histogram sub command but adds metadata in output for easy plotting,
also actually runs multi-threaded.
* gcp: K-mer GC Processor. Creates a matrix of the number of K-mers found given a GC
count and a K-mer count.
* comp: K-mer comparison tool. Creates a matrix of shared K-mers between two (or three)
sequence files.
* sect: SEquence Coverage estimator Tool. Estimates the coverage of each sequence in
a file using K-mers from another sequence file.
* cold: Given, reads and an assembly, calculates both the read and assembly K-mer
coverage along with GC% for each sequence in the assembly.
a file using K-mers from another sequence file.
* filter: Filtering tools. Contains tools for filtering k-mers and sequences based on
user-defined GC and coverage limits.
* plot: Plotting tools. Contains several plotting tools to visualise K-mer and compare
distributions.
Options:
-v [ --verbose ] Print extra information
--version Print version string
--help Produce help message
ラン
hist: histogram of k-mer occurrences.
> kat hist
$ kat hist
Kmer Analysis Toolkit (KAT) V2.4.2
Usage: kat hist [options] (<input>)+
Create an histogram of k-mer occurrences from the input.
Create an histogram with the number of k-mers having a given count, derived from the input, which can take the form of a single jellyfish hash, or one or more FastA or FastQ files. In bucket 'i' are tallied the k-mers which have a count 'c' satisfying 'low+i*inc <= c < low+(i+1)'. Buckets in the output are labeled by the low end point (low+i).
The last bucket in the output behaves as a catchall: it tallies all k-mers with a count greater or equal to the low end point of this bucket.
This tool is very similar to the "histo" tool in jellyfish itself. The primary difference being that the output contains metadata that make the histogram easier for the user to plot.
Options:
-o [ --output_prefix ] arg (="kat.hist") Path prefix for files generated by this program.
-t [ --threads ] arg (=1) The number of threads to use
-l [ --low ] arg (=1) Low count value of histogram
-h [ --high ] arg (=10000) High count value of histogram
-i [ --inc ] arg (=1) Increment for each bin
--5ptrim arg (=0) Ignore the first X bases from reads. If more that one file is provided you can specify different values for each file by
seperating with commas.
-N [ --non_canonical ] If counting fast(a/q), store explicit kmer as found. By default, we store 'canonical' k-mers, which means we count both strands.
-m [ --mer_len ] arg (=27) The kmer length to use in the kmer hashes. Larger values will provide more discriminating power between kmers but at the expense
of additional memory and lower coverage.
-H [ --hash_size ] arg (=100000000) If kmer counting is required for the input, then use this value as the hash size. If this hash size is not large enough for your
dataset then the default behaviour is to double the size of the hash and recount, which will increase runtime and memory usage.
-d [ --dump_hash ] Dumps any jellyfish hashes to disk that were produced during this run. Normally, this is not recommended, as hashes are slow to
load and will likely consume a significant amount of disk space.
-p [ --output_type ] arg (=png) The plot file type to create: png, ps, pdf.
-v [ --verbose ] Print extra information.
--help Produce help message.
k-merサイズ27でペアエンドのfastqを分析。横軸の最大値は10000とする。
kat hist -t 8 -l 1 -h 10000 -m 27 -v -o output sample1_*.fastq
頻度を示したテキストファイルとヒストグラムのグラフが出力される。下はシロイヌナズナのfastq分析結果。

gcp: Creates a matrix of the number of K-mers found given a GC count and a K-mer count
> kat gcp
$ kat gcp
Kmer Analysis Toolkit (KAT) V2.4.2
Usage: kat gcp (<input>)+
Compares GC content and K-mer coverage from the input.
This tool takes in either a single jellyfish hash or one or more FastA or FastQ input files and then counts the GC nucleotides for each distinct K-mer in the hash. For each GC count and K-mer coverage level, the number of distinct K-mers are counted and stored in a matrix. This matrix can be used to analyse biological content within the hash. For example, it can be used to distinguish legitimate content from contamination, or unexpected content.
Options:
-o [ --output_prefix ] arg (="kat-gcp") Path prefix for files generated by this program.
-t [ --threads ] arg (=1) The number of threads to use
-x [ --cvg_scale ] arg (=1) Number of bins for the gc data when creating the contamination matrix.
-y [ --cvg_bins ] arg (=1000) Number of bins for the cvg data when creating the contamination matrix.
--5ptrim arg (=0) Ignore the first X bases from reads. If more that one file is provided you can specify different values for each file by seperating
with commas.
-N [ --non_canonical ] If counting fast(a/q), store explicit kmer as found. By default, we store 'canonical' k-mers, which means we count both strands.
-m [ --mer_len ] arg (=27) The kmer length to use in the kmer hashes. Larger values will provide more discriminating power between kmers but at the expense of
additional memory and lower coverage.
-H [ --hash_size ] arg (=100000000) If kmer counting is required for the input, then use this value as the hash size. If this hash size is not large enough for your
dataset then the default behaviour is to double the size of the hash and recount, which will increase runtime and memory usage.
-d [ --dump_hash ] Dumps any jellyfish hashes to disk that were produced during this run. Normally, this is not recommended, as hashes are slow to load
and will likely consume a significant amount of disk space.
-p [ --output_type ] arg (=png) The plot file type to create: png, ps, pdf.
-v [ --verbose ] Print extra information.
--help Produce help message.
k-merサイズ27でペアエンドのfastqを分析。
kat gcp -t 10 -x 1 -y 10000 -m 27 -o gcc -v sample1_*.fastq
シロイヌナズナのシーケンスデータ分析結果。
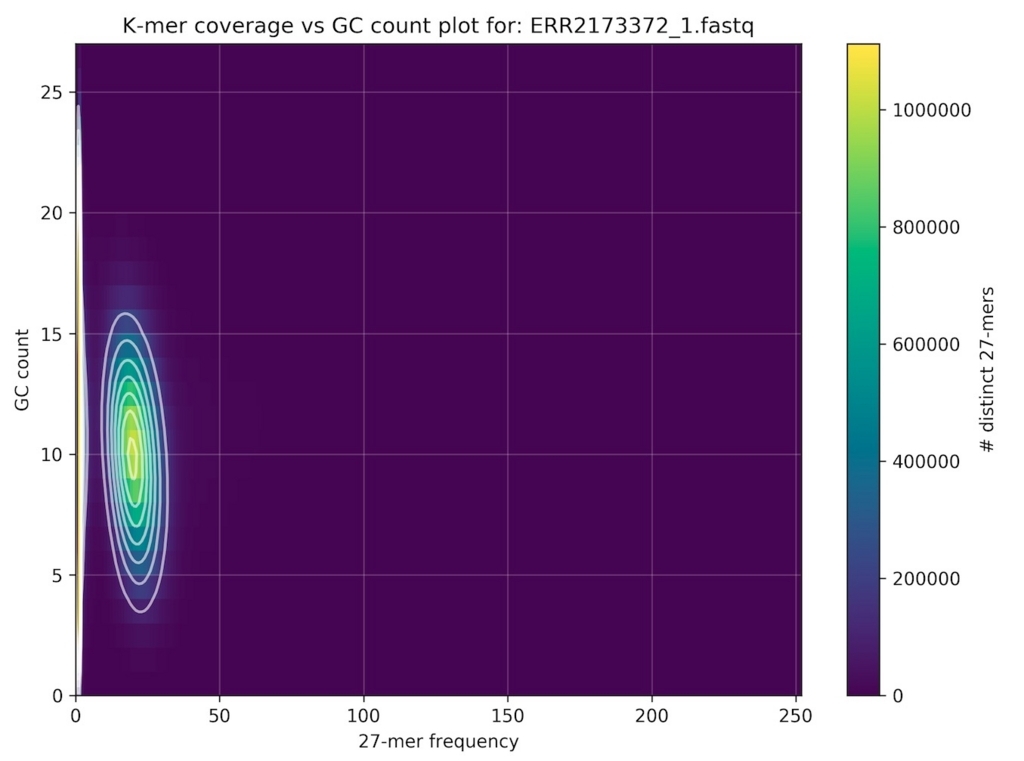
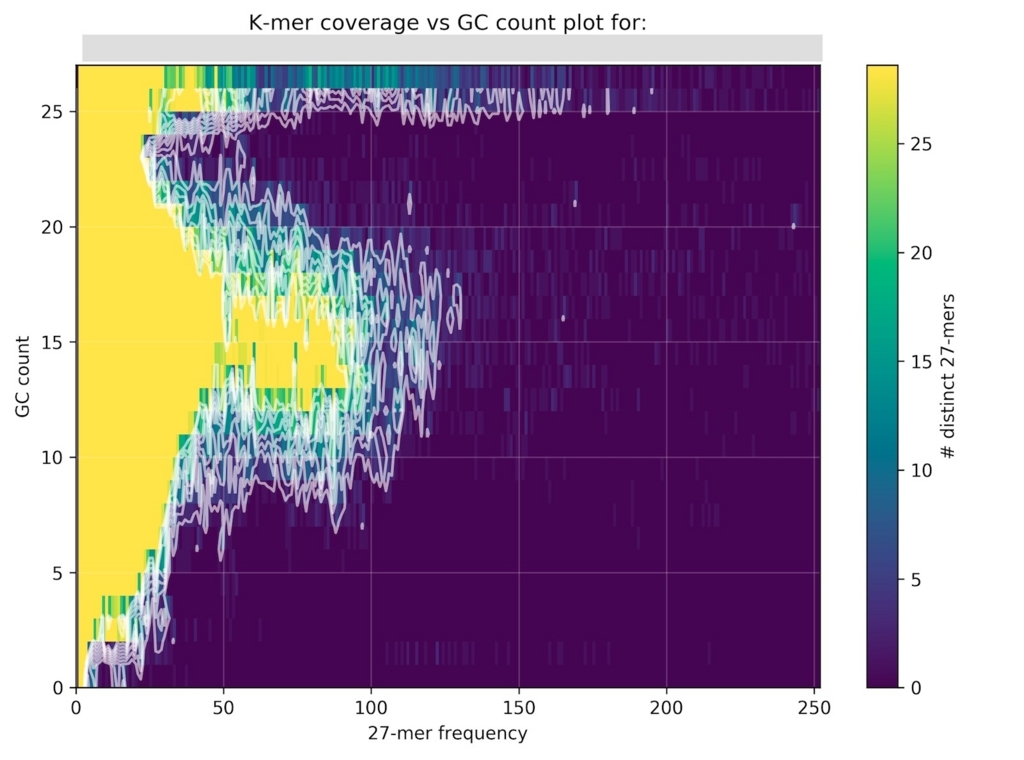
激しくコンタミがある バクテリアのシーケンスデータ分析結果。
comp: Compares jellyfish K-mer count hashes.
> kat comp
$ kat comp
Kmer Analysis Toolkit (KAT) V2.4.2
Usage: kat comp [options] <input_1> <input_2> [<input_3>]
Compares jellyfish K-mer count hashes.
There are two main use cases for this tool. The first is to compare K-mers from two K-mer hashes both representing K-mer counts for reads. The intersected output forms a matrix that can be used to show how related both spectra are via a density plot. The second use case is to compare K-mers generated from reads to those generated from an assembly, in this case the dataset for the reads must be provided first and the assembly second. This also produces a matrix containing the intersection of both spectra, but this is instead visualised via a stacked histogram.
There is also a third use case where K-mers from a third dataset as a filter, restricting the analysis to the K-mers present on that set. The manual contains more details on specific use cases.
Options:
-o [ --output_prefix ] arg (=kat-comp) Path prefix for files generated by this program.
-t [ --threads ] arg (=1) The number of threads to use.
-x [ --d1_scale ] arg (=1) Scaling factor for the first dataset - float multiplier
-y [ --d2_scale ] arg (=1) Scaling factor for the second dataset - float multiplier
-i [ --d1_bins ] arg (=1001) Number of bins for the first dataset. i.e. number of rows in the matrix
-j [ --d2_bins ] arg (=1001) Number of bins for the second dataset. i.e. number of rows in the matrix
--d1_5ptrim arg (=0) Ignore the first X bases from reads in dataset 1. If more that one file is provided for dataset 1 you can specify different values
for each file by seperating with commas.
--d2_5ptrim arg (=0) Ignore the first X bases from reads in dataset 2. If more that one file is provided for dataset 2 you can specify different values
for each file by seperating with commas.
-N [ --non_canonical_1 ] If counting fast(a/q) for input 1, store explicit kmer as found. By default, we store 'canonical' k-mers, which means we count both
strands.
-O [ --non_canonical_2 ] If counting fast(a/q) for input 2, store explicit kmer as found. By default, we store 'canonical' k-mers, which means we count both
strands.
-P [ --non_canonical_3 ] If counting fast(a/q) for input 3, store explicit kmer as found. By default, we store 'canonical' k-mers, which means we count both
strands.
-m [ --mer_len ] arg (=27) The kmer length to use in the kmer hashes. Larger values will provide more discriminating power between kmers but at the expense of
additional memory and lower coverage.
-H [ --hash_size_1 ] arg (=100000000) If kmer counting is required for input 1, then use this value as the hash size. If this hash size is not large enough for your
dataset then the default behaviour is to double the size of the hash and recount, which will increase runtime and memory usage.
-I [ --hash_size_2 ] arg (=100000000) If kmer counting is required for input 2, then use this value as the hash size. If this hash size is not large enough for your
dataset then the default behaviour is to double the size of the hash and recount, which will increase runtime and memory usage.
-J [ --hash_size_3 ] arg (=100000000) If kmer counting is required for input 3, then use this value as the hash size. If this hash size is not large enough for your
dataset then the default behaviour is to double the size of the hash and recount, which will increase runtime and memory usage.
-d [ --dump_hashes ] Dumps any jellyfish hashes to disk that were produced during this run. Normally, this is not recommended, as hashes are slow to load
and will likely consume a significant amount of disk space.
-g [ --disable_hash_grow ] By default jellyfish will double the size of the hash if it gets filled, and then attempt to recount. Setting this option to true,
disables automatic hash growing. If the hash gets filled an error is thrown. This option is useful if you are working with large
genomes, or have strict memory limits on your system.
-n [ --density_plot ] Makes a density plot. By default we create a spectra_cn plot.
-p [ --output_type ] arg (=png) The plot file type to create: png, ps, pdf.
-h [ --output_hists ] Whether or not to output histogram data and plots for input 1 and input 2
-v [ --verbose ] Print extra information.
--help Produce help message.
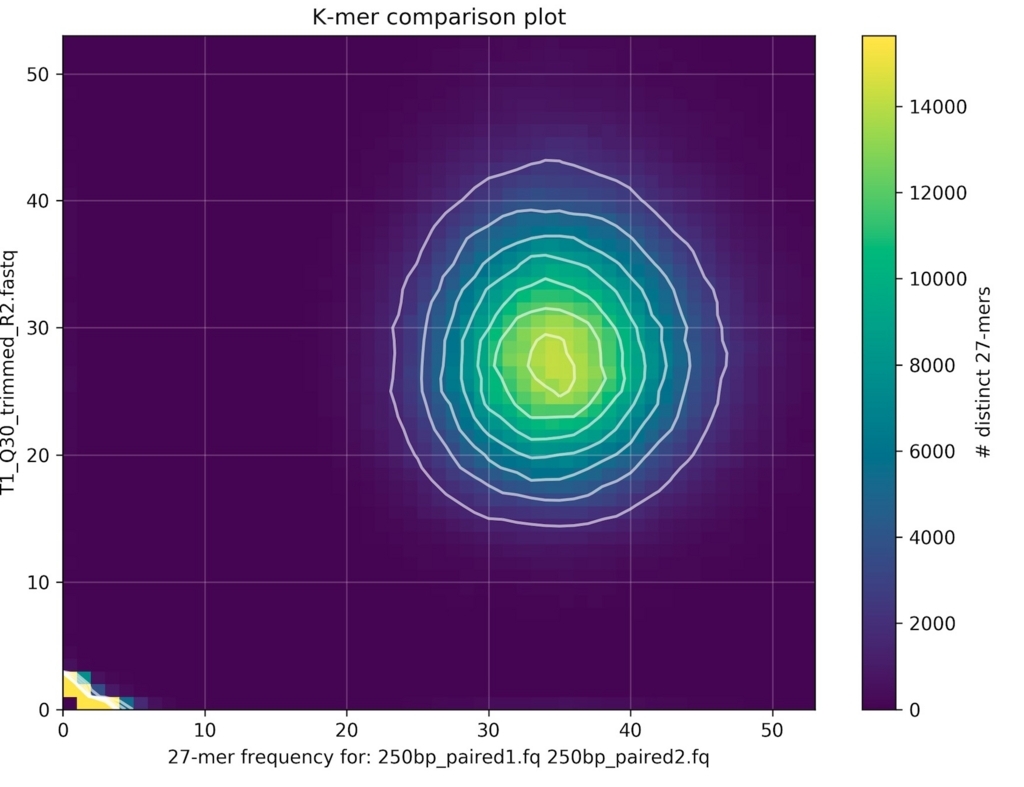
2サンプルのfastqを比較する。
kat comp -t 10 -v 'sample1_1.fq sample1_2.fq' 'sample2_1.fq sample2_2.fq'
kat-comp-main.mxとヒストグラムが出力される。ここではここでは同じゲノムのリアルデータ(PCR free)とシミュレーションデータを比較した。

plotコマンドで2サンプルのbiasを分析。density plot出力。
kat plot density kat-comp-main.mx
plotコマンドで2サンプルのbiasを分析。ヒストグラム出力。
kat plot spectra-mx -i kat-comp-main.mx
cold: Contig Length and Duplication analysis tool.
> kat cold
$ kat cold
Kmer Analysis Toolkit (KAT) V2.4.2
Usage: kat cold [options] <assembly> <reads>
COntig Length and Duplication analysis tool
Calculates median read k-mer coverage, assembly k-mer coverage and GC% across each sequence in the provided assembly. Then, assuming plotting is enabled, the results are converted into a scatter plot, where each point is colored according to a similar scheme used in spectra-cn plots, and sized according to its length. The y-axis representsmedian read K-mer coverage, and x-axis represents GC%.
The <assembly> should be a fasta file that is NOT gzipped compressed. The <reads> can be any number of <fasta/q> files, which CAN be gzipped compressed, or a pre-counted hash.
Options:
-o [ --output_prefix ] arg (="kat-cold") Path prefix for files generated by this program.
-x [ --gc_bins ] arg (=1001) Number of bins for the gc data when creating the contamination matrix.
-y [ --cvg_bins ] arg (=1001) Number of bins for the cvg data when creating the contamination matrix.
-t [ --threads ] arg (=1) The number of threads to use
--5ptrim arg (=0) Ignore the first X bases from reads. If more that one file is provided you can specify different values for each file by
seperating with commas.
-m [ --mer_len ] arg (=27) The kmer length to use in the kmer hashes. Larger values will provide more discriminating power between kmers but at the expense
of additional memory and lower coverage.
-H [ --hash_size ] arg (=100000000) If kmer counting is required, then use this value as the hash size for the reads. We assume the assembly should use half this
value. If this hash size is not large enough for your dataset then the default behaviour is to double the size of the hash and
recount, which will increase runtime and memory usage.
-d [ --dump_hashes ] Dumps any jellyfish hashes to disk that were produced during this run. Normally, this is not recommended, as hashes are slow to
load and will likely consume a significant amount of disk space.
-g [ --disable_hash_grow ] By default jellyfish will double the size of the hash if it gets filled, and then attempt to recount. Setting this option to true,
disables automatic hash growing. If the hash gets filled an error is thrown. This option is useful if you are working with large
genomes, or have strict memory limits on your system.
-p [ --output_type ] arg (=png) The plot file type to create: png, ps, pdf.
-v [ --verbose ] Print extra information.
--help Produce help message.
アセンブリしたfastaと、アセンブリに使ったfastqを指定する。
kat cold -t 10 -v assembly.fasta sample1_*.fastq
少しだけコンタミがある バクテリアのシーケンスデータと、そのspadesアセンブリFASTAの分析結果。

filter: K-mer filtering tools。kmerとseqがある。
> kat filter
$ kat filter
Kmer Analysis Toolkit (KAT) V2.4.2
Usage: kat filter <mode>
Filtering tools
First argument should be the filter mode you wish to use:
* kmer: Filters a jellyfish k-mer hash using user defined properties.
* seq: Filters sequences in a file based on k-mers in a given hash
Options:
-v [ --verbose ] Print extra information
--help Produce help message.
はじめにjellyfishなどでk-mer分析を行う。
jellyfish count -m 27 -s 10000000 -t 10sample1_*fq -o mer_counts.jf
k-mer 頻度10以上1000以下、k-merのGC率50-60%だけ抽出する。
kat filter kmer -t 10 -c 5 -d 1000 -g 50 -h 60 -o filtered mer_counts.jf
plot: Plotting tools.
> kat plot
$ kat plot
Kmer Analysis Toolkit (KAT) V2.4.2
Usage: kat plot <mode>
Create K-mer Plots
First argument should be the plot mode you wish to use:
* density: Creates a density plot from a matrix created with the "comp" tool or the
"GCP" tool. Typically this is used to compare two K-mer hashes produced
by different NGS reads, or to represent the kmer coverage vs GC count plots.
* profile: Creates a K-mer coverage plot for a single sequence. Takes in fasta
coverage output from the "sect" tool
* spectra-cn: Creates a stacked histogram using a matrix created with the "comp" tool.
Typically this is used to compare a jellyfish hash produced from a read set
to a jellyfish hash produced from an assembly. The plot shows the amount of
distinct K-mers absent, as well as the copy number variation present within
the assembly.
* spectra-hist: Creates a K-mer spectra plot for a set of K-mer histograms produced either
by jellyfish-histo or kat-histo.
* spectra-mx: Creates a K-mer spectra plot for a set of K-mer histograms that are derived
from selected rows or columns in a matrix produced by the "comp".
* cold: Takes in a stats file produced by "cold" and produces a scatter plot of
median read K-mer coverage vs GC% for each contig in the assembly, with
point's size being adjusted by sequence length
Options:
-v [ --verbose ] Print extra information
--help Produce help message.
plotはcompコマンドでの例のように、他のコマンドと組み合わせて使う。
この他、カバレッジを推定するsectがある。
KATマニュアルのwalkthroughセクションを読むと、サンブルのバッチエフェクト、シーケンスバイアスの調べ方などを学べます。
http://kat.readthedocs.io/en/latest/walkthrough.html#comparing-r1-v-r2-in-an-illumina-pe-dataset
追記
ゲノムサイズ推定
引用
KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies
Daniel Mapleson, Gonzalo Garcia Accinelli, George Kettleborough, Jonathan Wright, and Bernardo J. Clavijo
Bioinformatics. 2017 Feb 15; 33(4): 574–576.