2020 5/25 docker link追加、ヘルプ追加、分かりにくい文章の修正
murasakiは複数ゲノムの相同性ある領域の探索を高速に行うツールで、GMVはその比較結果を見るためのビューアソフトである。領域によってカラフルな色がつくので、ゲノムリアレンジメントなどの構造変化をわかりやすく示すことができる。
公式サイト
murasaki genomemurasaki genome
murasakiのインストールはこの方のブログを参考にしてください。
2020 5/26 追記
#dockerhub(link)
docker pull biocontainers/murasaki:v1.68.6-8b1-deb_cv1
> docker run --rm -it biocontainers/murasaki:v1.68.6-8b1-deb_cv1 murasaki -h
$ docker run --rm -it biocontainers/murasaki:v1.68.6-8b1-deb_cv1 murasaki -h
Usage: murasaki -p<patternSpec> [options] seq1 [seq2 [seq3 ... ]]
Options:
--pattern|-p = seed pattern (eg. 11101001010011011).
using the format [<w>:<l>] automatically generates a
random pattern of weight <w> and length <l>
--directory|-d = output directory (default: output)
--name|-n = alignment name (default: test)
--quickhash|-q = specify a hashing function:
0 - adaptive with S-boxes
1 - don't pack bits to make hash (use first word only)
2 - naively use the first hashbits worth of pattern
3 - adaptivevely find a good hash (default)
**experimental CryptoPP hashes**
4 - MD5
5 - SHA1
6 - Whirlpool
7 - CRC-32
8 - Adler-32
--hashbits|-b = use n bit hashes (for n's of 1 to WORDSIZE. default 26)
--hashtype|-t = select hash table data structure to use:
OpenHash - open sub-word packing of hashbits
EcoHash - chained sub-word packing of hashbits (default)
ArrayHash - malloc/realloc (fast but fragmenty)
MSetHash - memory exorbanant, almost pointless.
--probing = 0 - linear, 1 - quadratic (default)
--hitfilter|-h = minimum number of hits to be outputted as an anchor
(default 1)
--histogram|-H = histogram computation level: (-H alone implies -H1)
0 - no histogram (default)
1 - basic bucketsize/bucketcount histogram data
2 - bucket-based scores to anchors.detils
3 - perbucket count data
4 - perbucket + perpattern count data
--repeatmask|-r= skip repeat masked data (ie: lowercase atgc)
--seedfilter|-f= skip seeds that occur more than N times
--hashfilter|-m= like --seedfilter but works on hash keys instead of
seeds. May cause some collateral damage to otherwise
unique seeds, but it's faster. Also non-sequence-specific
so more like at best 1/N the tolerance of seedfilter.
--rseed|-s = random number seed for non-deterministic algorithms
(ie: the adative hash-finding). If you're doing any
performance comparisons, it's probably imperative that you
use the same seed for each run of the same settings.
Default is obtained from time() (ie: seconds since 1970).
--skipfwd|-F = Skip forward facing matches
--skiprev|-R = Skip reverse facing matches
--skip1to1|-1 = Skip matches along the 1:1 line (good for comparing to self)
--hashonly|-Q = Hash Only. no anchors. just statistics.
--hashskip|-S = Hashes every n bases. (Default is 1. ie all)
Not supplying any argument increments the skip amount by 1.
--hashCache|-c = Caches hash tables in the directory. (default: cache/)
--join|-j = Join anchors within n bases of eachother (default: 0)
Specifying a negative n implies -n*patternLength
--bitscore|-B = toggles compututation of a bitscore for all anchors
(default is on)
--memory|-M = set the target amount of total memory
(either in gb or as % total memory)
--seedterms|-T = toggles retention of seed terms (defaults to off)
(these are necessary for computing TF-IDF scores)
--sectime|-e = always display times in seconds
--repeatmap|-i = toggles keeping of a repeat map when --mergefilter
is used (defaults to yes).
--mergefilter|-Y = filter out matches which would would cause more than N
many anchors to be generated from 1 seed (default -Y100).
Use -Y0 to disable.
--scorefilter = set a minimum ungapped score for seeds
--tfidf|-k = perform accurate tfidf scoring from within murasaki
(requires extra memory at anchor generation time)
--reverseotf|-o = generate reverse complement on the fly (defaults to on)
--rifts|-/ = allow anchors to skip N sequences (default 0)
--islands|-% = same as --rifts=S-N (where S is number of seqs)
--fuzzyextend|-z = enable (default) or disable fuzzy extension of hits
--fuzzyextendlosslimit|-Z = set the cutoff at which to stop extending
fuzzy hits (ie. the BLAST X parameter).
--gappedanchors = use gapped (yes) or ungapped (no (default)) anchors.
--scorebyminimumpair = do anchor scoring by minimum pair when appropriate
(default). Alternative is mean (somewhat illogical, but
theoretically faster).
--binaryseq = enable (default) or disable binary sequence read/write
Adaptive has function related:
--hasherFairEntropy = use more balanced entropy estimation (default: yes)
--hasherCorrelationAdjust = adjust entropy estimates for nearby sources
assuming some correlation (default: yes)
--hasherTargetGACycles = GA cycle cutoff
--hasherEntropyAgro = how aggressive to be about pursuing maximum
entropy hash functions (takes a real. default is 1).
...and of course
--verbose|-v = increases verbosity
--version|-V = prints version information and quits
--help|-? = prints this help message and quits
Platform information:
Wordsize: 64 bits
sizeof(word): 8 bytes
Total Memory: 22.97 GB
Available Memory: 23.93 GB (104.20%)
Murasaki version 1.68.6 (LARGESEQ, CRYPTOPP)
ラン
ゲノム・プラスミドのfastaかgenbakファイルを指定する。
3つのgbkファイルの比較。
./murasaki -p 19:26 -d output -n sample_name 7942.gbk GT-S.gbk 7002.gbk Leptolyngbya_dg5.gbk Nostoc_sp.PCC7120.gbk
#docker imageを使うなら
docker run --rm -itv $PWD:/data/ -w /data biocontainers/murasaki:v1.68.6-8b1-deb_cv1 murasaki A.fasta B.fasta C.fasta -p 19:26 -d output
出力ディレクトリoutputをGNVに読み込ませて描画すると、下のような図を出力できる。
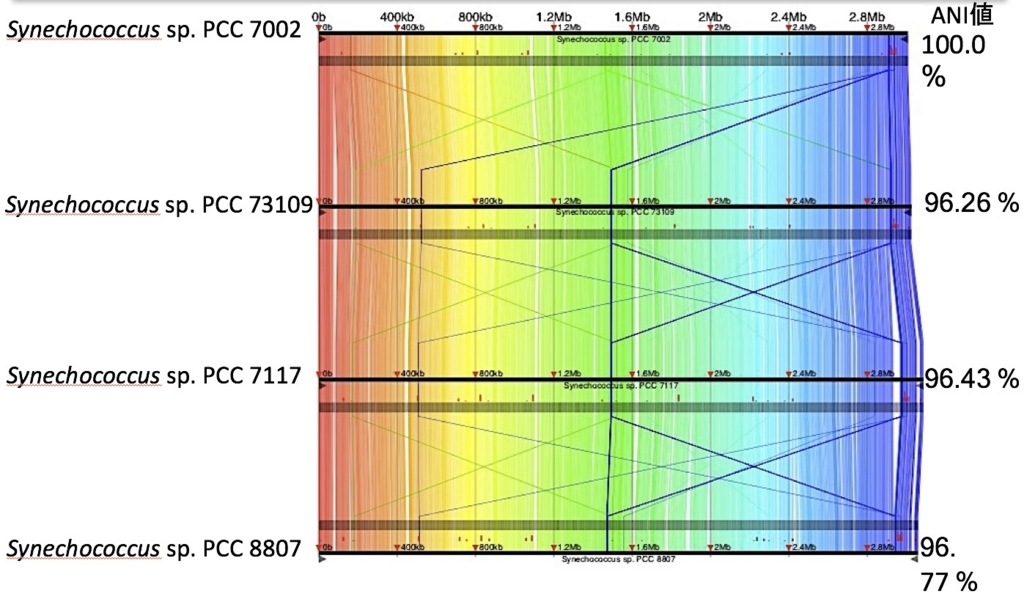
4種の非常に近縁なシアノバクテリアを比較。右端にはANI値を載せた。
ランさせた時の様子はこのような感じになる。
動画では、genbankファイルを2つ選択してmurasakiで相同性を調べ、出力されたフォルダをGMVで表示している。
Yasunori Osana @ University of the Ryukyus
こちらも貼っておきます。
Yasunori Osana @ University of the Ryukyus
引用
Murasaki: a fast, parallelizable algorithm to find anchors from multiple genomes
Popendorf K, Tsuyoshi H, Osana Y, Sakakibara Y
PLoS One. 2010 Sep 24;5(9):e12651