2021 2/15 追記
anvi'o パンゲノミックワークフローは、3 つの主要なステップで構成されている。
1、anvi-gen-genomes-storage による anvi'o ゲノムストレージ生成
2、anvi-pan-genomeによるパンデータベース生成
3、anvi-display-pan(ゲノムストレージと パンデータベースが必要)を使用して結果を表示
そして、インタラクティブなインターフェースを使用して遺伝子クラスタをコレクションにまとめたりサマリーレポートを作成したりできる。チュートリアルに沿って使い方を見ていく。
インストール
公式dockerイメージを使ってubuntu18.04LTSマシンでテストした。
本体 Github
#依存が多いので、condaだと依存チェックに異常な時間がかかる。動かすだけならdockerが楽。
#latest (v6)
docker pull meren/anvio:latest
インストールチェック
> anvi-self-test --suite pangenomics
起動
docker run --rm -it -v `pwd`:`pwd` -w `pwd` -p 8080:8080 meren/anvio:latest
> anvi-migrate -h
> anvi-migrate -h
usage: anvi-migrate [-h] [--just-do-it] [-t VERSION] DATABASE [DATABASE ...]
Migrate an anvi'o database or config file to a newer version.
positional arguments:
DATABASE Anvi'o database or config file for migration
optional arguments:
-h, --help show this help message and exit
--just-do-it Do not bother me with warnings
-t VERSION, --target-version VERSION
Anvi'o will stop upgrading your database when it
reaches to this version.
:: anvi'o v6.2 ::
> anvi-gen-genomes-storage -h
> anvi-gen-genomes-storage -h
usage: anvi-gen-genomes-storage [-h] [-e FILE_PATH] [-i FILE_PATH]
[--gene-caller GENE-CALLER] -o GENOMES_STORAGE
Create a genome storage from internal or external genomes for a pan genome
analysis.
optional arguments:
-h, --help show this help message and exit
EXTERNAL GENOMES:
External genomes listed as anvi'o contigs databases. As in, you have one
or more genomes say from NCBI you want to work with, and you created an
anvi'o contigs database for each one of them.
-e FILE_PATH, --external-genomes FILE_PATH
A two-column TAB-delimited flat text file that lists
anvi'o contigs databases. The first item in the header
line should read 'name', and the second should read
'contigs_db_path'. Each line in the file should
describe a single entry, where the first column is the
name of the genome (or MAG), and the second column is
the anvi'o contigs database generated for this genome.
INTERNAL GENOMES:
Genome bins stored in an anvi'o profile databases as collections.
-i FILE_PATH, --internal-genomes FILE_PATH
A five-column TAB-delimited flat text file. The header
line must contain these columns: 'name', 'bin_id',
'collection_id', 'profile_db_path', 'contigs_db_path'.
Each line should list a single entry, where 'name' can
be any name to describe the anvi'o bin identified as
'bin_id' that is stored in a collection.
PRO STUFF:
Things you may not have to change. But you never know (unless you read the
help).
--gene-caller GENE-CALLER
The gene caller to utilize. Anvi'o supports multiple
gene callers, and some operations (including this one)
requires an explicit mentioning of which one to use.
The default is 'prodigal', but it will not be enough
if you if you were a rebel and have used `--external-
gene-callers` or something.
OUTPUT:
Give it a nice name. Must end with '-GENOMES.db'. This is primarily due to
the fact that there are other .db files used throughout anvi'o and it
would be better to distinguish this very special file from them.
-o GENOMES_STORAGE, --output-file GENOMES_STORAGE
File path to store results.
:: anvi'o v6.2 ::
> anvi-pan-genome -h
> anvi-pan-genome -h
WARNING
===============================================
If you publish results from this workflow, please do not forget to cite DIAMOND
(doi:10.1038/nmeth.3176), unless you use it with --use-ncbi-blast flag, and MCL
(http://micans.org/mcl/ and doi:10.1007/978-1-61779-361-5_15)
usage: anvi-pan-genome [-h] -g GENOMES_STORAGE [-G GENOME_NAMES]
[--skip-alignments] [--skip-homogeneity]
[--quick-homogeneity] [--align-with ALIGNER]
[--exclude-partial-gene-calls] [--use-ncbi-blast]
[--minbit MINBIT] [--mcl-inflation INFLATION]
[--min-occurrence NUM_OCCURRENCE]
[--min-percent-identity PERCENT] [--sensitive]
[-n PROJECT_NAME] [--description TEXT_FILE]
[-o PAN_DB_DIR] [-W] [-T NUM_THREADS]
[--skip-hierarchical-clustering]
[--enforce-hierarchical-clustering]
[--distance DISTANCE_METRIC] [--linkage LINKAGE_METHOD]
A DIAMOND and MCL-based anvi'o workflow for pangenomics. You provide genomes
from anywhere (whether they are external genomes, or anvi'o genome bins in
collections), and it gives you back a pangenome analysis.
optional arguments:
-h, --help show this help message and exit
GENOMES:
The very fancy genomes storage file. This file is generated by the program
`anvi-genomes-storage`. Please see the online tutorial on pangenomic
workflow if you don't know how to generate one.
-g GENOMES_STORAGE, --genomes-storage GENOMES_STORAGE
Anvi'o genomes storage file
-G GENOME_NAMES, --genome-names GENOME_NAMES
Genome names to 'focus'. You can use this parameter to
limit the genomes included in your analysis. You can
provide these names as a comma-separated list of
names, or you can put them in a file, where you have a
single genome name in each line, and provide the file
path.
PARAMETERS:
Important stuff Tom never pays attention (but you should).
--skip-alignments By default, anvi'o attempts to align amino acid
sequences in each gene cluster using multiple sequnce
alignment via muscle. You can use this flag to skip
that step and be upset later.
--skip-homogeneity By default, anvi'o attempts to calculate homogeneity
values for every gene cluster, given that they are
aligned. You can use this flag to have anvi'o skip
homogeneity calculations. Anvi'o will ignore this flag
if you decide to skip alignments
--quick-homogeneity By default, anvi'o will use a homogeneity algorithm
that checks for horizontal and vertical geometric
homogeneity (along with functional). With this flag,
you can tell anvi'o to skip horizontal geometric
homogeneity calculations. It will be less accurate but
quicker. Anvi'o will ignore this flag if you skip
homogeneity calculations or alignments all together.
--align-with ALIGNER The multiple sequence alignment program to use when
multiple sequence alignment is necessary. To see all
available options, use the flag `--list-aligners`.
--exclude-partial-gene-calls
By default, anvi'o includes all partial gene calls
from the analysis, which, in some cases, may inflate
the number of gene clusters identified and introduce
extra heterogeneity within those gene clusters. Using
this flag, you can request anvi'o to exclude partial
gene calls from the analysis (whether a gene call is
partial or not is an information that comes directly
from the gene caller used to identify genes during the
generation of the contigs database).
--use-ncbi-blast This program uses DIAMOND by default, however, if you
like, you can use good ol' blastp from NCBI instead.
--minbit MINBIT The minimum minbit value. The minbit heuristic
provides a mean to set a to eliminate weak matches
between two amino acid sequences. We learned it from
ITEP (Benedict MN et al, doi:10.1186/1471-2164-15-8),
which is a comprehensive analysis workflow for
pangenomes, and decided to use it in the anvi'o
pangenomic workflow, as well. Briefly, If you have two
amino acid sequences, 'A' and 'B', the minbit is
defined as 'BITSCORE(A, B) / MIN(BITSCORE(A, A),
BITSCORE(B, B))'. So the minbit score between two
sequences goes to 1 if they are very similar over the
entire length of the 'shorter' amino acid sequence,
and goes to 0 if (1) they match over a very short
stretch compared even to the length of the shorter
amino acid sequence or (2) the match betwen sequence
identity is low. The default is 0.5.
--mcl-inflation INFLATION
MCL inflation parameter, that defines the sensitivity
of the algorithm during the identification of the gene
clusters. More information on this parameter and it's
effect on cluster granularity is here:
(http://micans.org/mcl/man/mclfaq.html#faq7.2). The
default is 2.
--min-occurrence NUM_OCCURRENCE
Do you not want singletons?\ You don't? Well, this
parameter will help you get rid of them (along with
doubletons, if you want). Anvi'o will remove gene
clusters that occur less than the number you set using
this parameter from the analysis. The default is 1,
which means everything will be kept. If you want to
remove singletons, set it to 2, if you want to remove
doubletons as well, set it to 3, and so on.
--min-percent-identity PERCENT
Minimum percent identity between the two amino acid
sequences for them to have an edge for MCL analysis.
This value will be used to filter hits from Diamond
search results. Because percent identity is not a
predictor of a good match (since it does not
communicate many other important factors such as the
alignment length between the two sequences and its
proportion to the entire length of those involved), we
suggest you rely on 'minbit' parameter. But you know
what? Maybe you shouldn't listen to anyone, and
experiment on your own! The default is 0 percent.
--sensitive DIAMOND sensitivity. With this flag you can instruct
DIAMOND to be 'sensitive', rather than 'fast' during
the search. It is likely the search will take
remarkably longer. But, hey, if you are doing it for
your final analysis, maybe it should take longer and
be more accurate. This flag is only relevant if you
are running DIAMOND.
OTHERS:
Sweet parameters of convenience.
-n PROJECT_NAME, --project-name PROJECT_NAME
Name of the project. Please choose a short but
descriptive name (so anvi'o can use it whenever she
needs to name an output file, or add a new table in a
database, or name her first born).
--description TEXT_FILE
A plain text file that contains some description about
the project. You can use Markdwon syntax. The
description text will be rendered and shown in all
relevant interfaces, including the anvi'o interactive
interface, or anvi'o summary outputs.
-o PAN_DB_DIR, --output-dir PAN_DB_DIR
Directory path for output files
-W, --overwrite-output-destinations
Overwrite if the output files and/or directories
exist.
-T NUM_THREADS, --num-threads NUM_THREADS
Maximum number of threads to use for multithreading
whenever possible. Very conservatively, the default is
1. It is a good idea to not exceed the number of CPUs
/ cores on your system. Plus, please be careful with
this option if you are running your commands on a SGE
--if you are clusterizing your runs, and asking for
multiple threads to use, you may deplete your
resources very fast.
ORGANIZING GENE CLUSTERs:
These are stuff that will change the clustering dendrogram of your gene
clusters.
--skip-hierarchical-clustering
Anvi'o attempts to generate a hierarchical clustering
of your gene clusters once it identifies them so you
can use `anvi-display-pan` to play with it. But if you
want to skip this step, this is your flag.
--enforce-hierarchical-clustering
If you want anvi'o to try to generate a hierarchical
clustering of your gene clusters even if the number of
gene clusters exceeds its suggested limit for
hierarchical clustering, you can use this flag to
enforce it. Are you are a rebel of some sorts? Or did
computers made you upset? Express your anger towards
machine using this flag.
--distance DISTANCE_METRIC
The distance metric for the clustering of gene
clusters. If you do not use this flag, the default
distance metric will be used for each clustering
configuration which is "euclidean".
--linkage LINKAGE_METHOD
The same story with the `--distance`, except, the
system default for this one is ward.
:: anvi'o v6.2 ::
>anvi-display-pan -h
> anvi-display-pan -h
usage: anvi-display-pan [-h] -p PAN_DB [-g GENOMES_STORAGE] [-d VIEW_DATA]
[-t NEWICK] [-V ADDITIONAL_VIEW]
[-A ADDITIONAL_LAYERS] [--view NAME] [--title NAME]
[--state-autoload NAME] [--collection-autoload NAME]
[--export-svg FILE_PATH] [--skip-init-functions]
[--dry-run] [--skip-auto-ordering] [-I IP_ADDR]
[-P INT] [--browser-path PATH] [--read-only]
[--server-only] [--password-protected]
[--user-server-shutdown]
Start an anvi'o server to display a pan-genome
optional arguments:
-h, --help show this help message and exit
INPUT FILES:
Input files from the pangenome analysis.
-p PAN_DB, --pan-db PAN_DB
Anvi'o pan database
-g GENOMES_STORAGE, --genomes-storage GENOMES_STORAGE
Anvi'o genomes storage file
OPTIONAL INPUTS:
Where the yay factor becomes a reality.
-d VIEW_DATA, --view-data VIEW_DATA
A TAB-delimited file for view data
-t NEWICK, --tree NEWICK
NEWICK formatted tree structure
ADDITIONAL STUFF:
Parameters to provide additional layers, views, or layer data.
-V ADDITIONAL_VIEW, --additional-view ADDITIONAL_VIEW
A TAB-delimited file for an additional view to be used
in the interface. This file should contain all split
names, and values for each of them in all samples.
Each column in this file must correspond to a sample
name. Content of this file will be called 'user_view',
which will be available as a new item in the 'views'
combo box in the interface
-A ADDITIONAL_LAYERS, --additional-layers ADDITIONAL_LAYERS
A TAB-delimited file for additional layers for splits.
The first column of this file must be split names, and
the remaining columns should be unique attributes. The
file does not need to contain all split names, or
values for each split in every column. Anvi'o will try
to deal with missing data nicely. Each column in this
file will be visualized as a new layer in the tree.
VISUALS RELATED:
Parameters that give access to various adjustements regarding the
interface.
--view NAME Start the interface with a pre-selected view. To see a
list of available views, use --show-views flag.
--title NAME Title for the interface. If you are working with a
RUNINFO dict, the title will be determined based on
information stored in that file. Regardless, you can
override that value using this parameter.
--state-autoload NAME
Automatically load previous saved state and draw tree.
To see a list of available states, use --show-states
flag.
--collection-autoload NAME
Automatically load a collection and draw tree. To see
a list of available collections, use --list-
collections flag.
--export-svg FILE_PATH
The SVG output file path.
SWEET PARAMS OF CONVENIENCE:
Parameters and flags that are not quite essential (but nice to have).
--skip-init-functions
When declared, function calls for genes will not be
initialized (therefore will be missing from all
relevant interfaces or output files). The use of this
flag may reduce the memory fingerprint and processing
time for large datasets.
--dry-run Don't do anything real. Test everything, and stop
right before wherever the developer said 'well, this
is enough testing', and decided to print out results.
--skip-auto-ordering When declared, the attempt to include automatically
generated orders of items based on additional data is
skipped. In case those buggers cause issues with your
data, and you still want to see your stuff and deal
with the other issue maybe later.
SERVER CONFIGURATION:
For power users.
-I IP_ADDR, --ip-address IP_ADDR
IP address for the HTTP server. The default ip address
(0.0.0.0) should work just fine for most.
-P INT, --port-number INT
Port number to use for anvi'o services. If nothing is
declared, anvi'o will try to find a suitable port
number, starting from the default port number, 8080.
--browser-path PATH By default, anvi'o will use your default browser to
launch the interactive interface. If you would like to
use something else than your system default, you can
provide a full path for an alternative browser using
this parameter, and hope for the best. For instance we
are using this parameter to call Google's experimental
browser, Canary, which performs better with demanding
visualizations.
--read-only When the interactive interface is started with this
flag, all 'database write' operations will be
disabled.
--server-only The default behavior is to start the local server, and
fire up a browser that connects to the server. If you
have other plans, and want to start the server without
calling the browser, this is the flag you need.
--password-protected If this flag is set, command line tool will ask you to
enter a password and interactive interface will be
only accessible after entering same password. This
option is recommended for shared machines like
clusters or shared networks where computers are not
isolated.
--user-server-shutdown
Allow users to shutdown an anvi'server via web
interface.
:: anvi'o v6.2 ::
> anvi-split -h
> anvi-split -h
usage: anvi-split [-h] -p PAN_OR_PROFILE_DB [-c CONTIGS_DB]
[-g GENOMES_STORAGE] [--skip-variability-tables]
[--compress-auxiliary-data] [-C COLLECTION_NAME]
[-b BIN_NAME] [-o DIR_PATH] [--list-collections]
[--skip-hierarchical-clustering]
[--enforce-hierarchical-clustering]
[--distance DISTANCE_METRIC] [--linkage LINKAGE_METHOD]
Split an anvi'o pan or profile database into smaller, self-contained pieces.
This is usually great when you want to share a subset of an anvi'o project.
You give this guy your databases, and a collection id, and it gives you back
directories of individual projects for each bin that can be treated as self-
contained smaller anvi'o projects. We know you don't read this far into these
help menus, but please remember: you will either need to provide a profile &
contigs database pair, or a pan & genomes storage pair. The rest will be taken
care of. Magic.
optional arguments:
-h, --help show this help message and exit
DATABASES:
You will either provide a PROFILE/CONTIGS or a PAN/GENOMES STORAGE pair
here.
-p PAN_OR_PROFILE_DB, --pan-or-profile-db PAN_OR_PROFILE_DB
Anvi'o pan or profile database (and even genes
database in appropriate contexts).
-c CONTIGS_DB, --contigs-db CONTIGS_DB
Anvi'o contigs database generated by 'anvi-gen-
contigs'
-g GENOMES_STORAGE, --genomes-storage GENOMES_STORAGE
Anvi'o genomes storage file
PROFILE/CONTIGS OPTIONS:
Some options that are specific to this only.
--skip-variability-tables
Processing variability tables in profile database
might take a very long time. With this flag you will
be asking anvi'o to skip them.
--compress-auxiliary-data
When declared, the auxiliary data file in the
resulting output will be compressed. This saves space,
but it takes long. Also, if you are planning to
compress the entire later using GZIP, it is even
useless to do. But you are the boss!
COLLECTION:
You should provide a valid collection name. If you do not provide bin
names, the program will generate an output for each bin in your collection
separately.
-C COLLECTION_NAME, --collection-name COLLECTION_NAME
Collection name.
-b BIN_NAME, --bin-id BIN_NAME
Bin name you are interested in.
OUTPUT:
Where do we want the resulting split profiles to be stored.
-o DIR_PATH, --output-dir DIR_PATH
Directory path for output files
EXTRAS:
Stuff that you rarely need, but you really really need when the time
comes. Following parameters will aply to each of the resulting anvi'o
profile that will be split from the mother anvi'o profile.
--list-collections Show available collections and exit.
--skip-hierarchical-clustering
If you are not planning to use the interactive
interface (or if you have other means to add a tree of
contigs in the database) you may skip the step where
hierarchical clustering of your items are preformed
based on default clustering recipes matching to your
database type.
--enforce-hierarchical-clustering
If you have more than 25,000 splits in your merged
profile, anvi-merge will automatically skip the
hierarchical clustering of splits (by setting --skip-
hierarchical-clustering flag on). This is due to the
fact that computational time required for hierarchical
clustering increases exponentially with the number of
items being clustered. Based on our experience we
decided that 25,000 splits is about the maximum we
should try. However, this is not a theoretical limit,
and you can overwrite this heuristic by using this
flag, which would tell anvi'o to attempt to cluster
splits regardless.
--distance DISTANCE_METRIC
The distance metric for the hierarchical clustering.
If you do not use this flag, the default distance
metric will be used for each clustering configuration
which is "euclidean".
--linkage LINKAGE_METHOD
The same story with the `--distance`, except, the
system default for this one is ward.
:: anvi'o v6.2 ::
ラン
チュートリアルに従って進める。
anvi'oのメタゲノムビニングFASTA(internal genome)、ユーザーが用意したFASTA(external genome)を利用できる。
1、データのダウンロード。チュートリアルが使っているProchlorococcusのデータになる。
wget https://ndownloader.figshare.com/files/11857577 -O Prochlorococcus_31_genomes.tar.gz
tar -zxvf Prochlorococcus_31_genomes.tar.gz
cd Prochlorococcus_31_genomes

各ゲノムの.dbファイルがダンロードされる。自分のデータを利用する場合、はじめにanvi-gen-contigs-databaseコマンドを使ってゲノム毎にデータベースファイルを作成してください。
2、起動と.dbの読み込み
#lauch
docker run --rm -it -v `pwd`:`pwd` -w `pwd` -p 8080:8080 meren/anvio:latest
#h5pyがないと怒られたら導入
pip install h5py
#すでにある.dbを更新
anvi-migrate *.db
更新された.dbファイルがそのままカレントに出力される。
3、 全ゲノムの.dbを出力。任意で各ゲノムの追加情報を含むTAB区切りファイル(external-genomes.txt)を指定する。
external-genomes.txtの中身
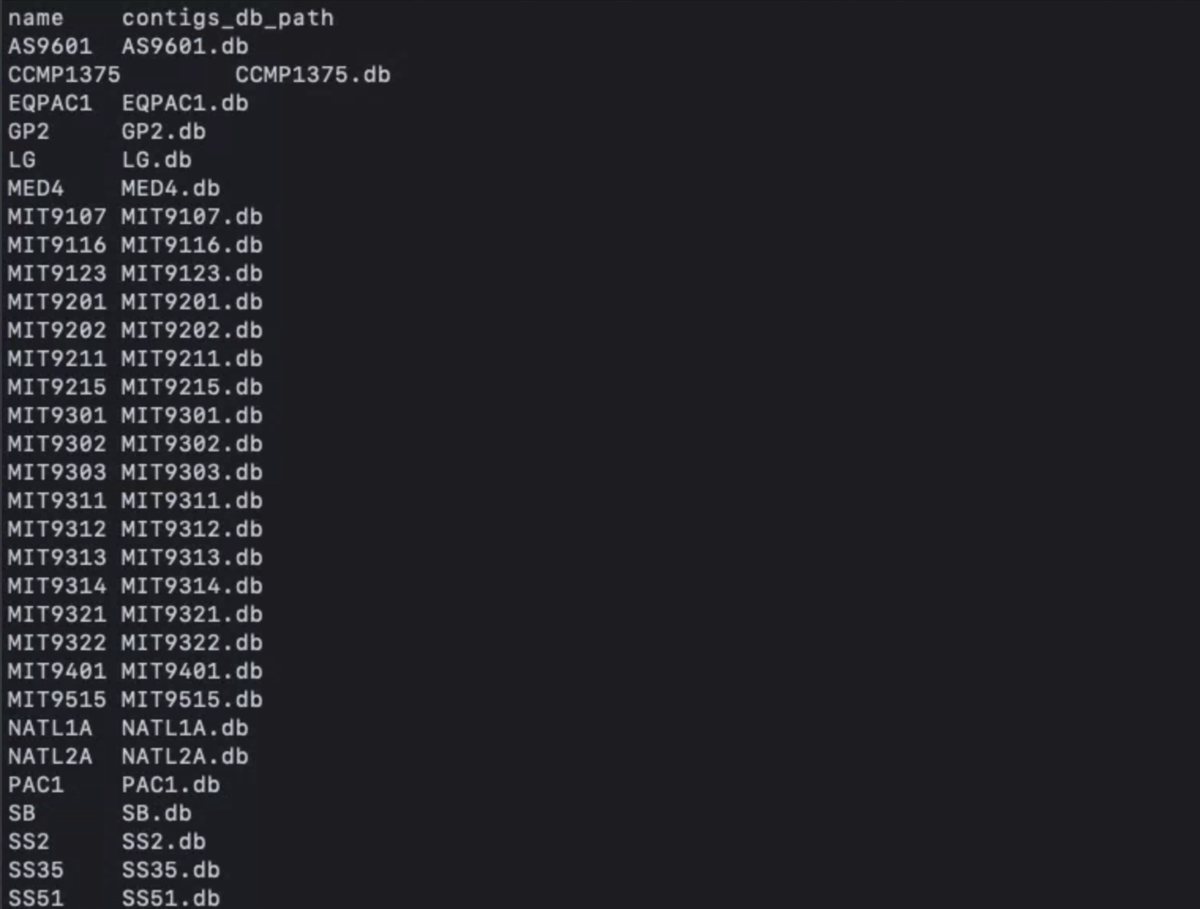
自分のデータを使う場合、タブ区切りで
name contigs_db_path
genome1 genome1.db
genome2 genome2.db
genome3 genome3.db
.
.
というファイルを用意する。
テキストを用意したら実行する。
anvi-gen-genomes-storage -e external-genomes.txt -o PROCHLORO-GENOMES.db
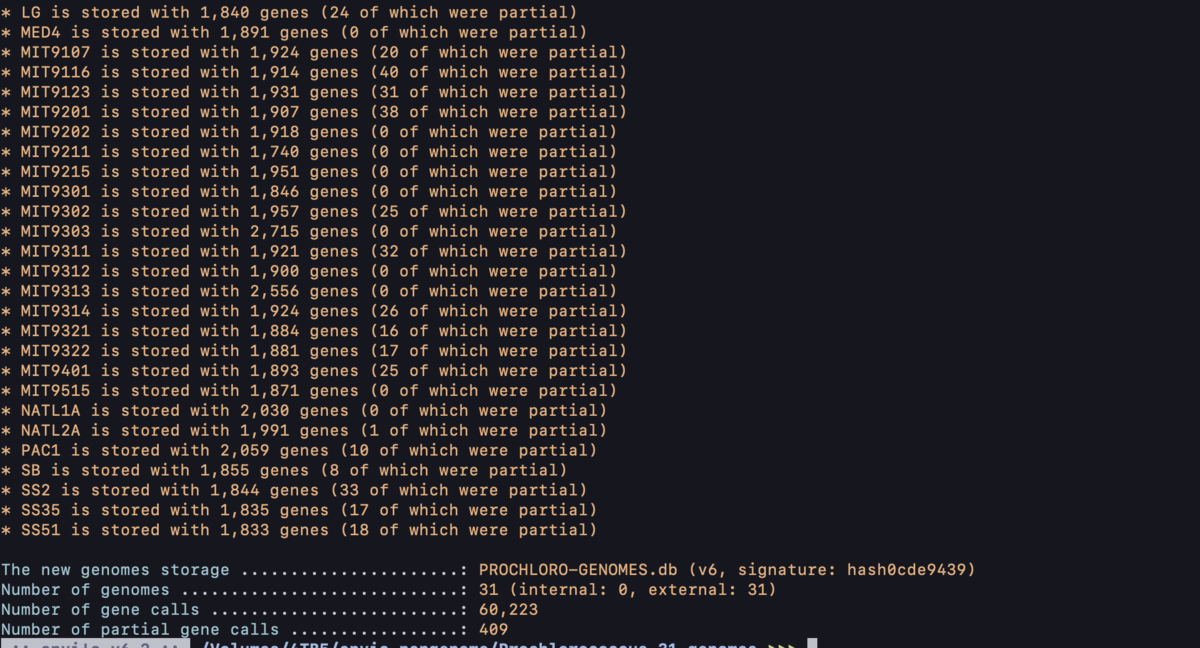
指定したPROCHLORO-GENOMES.dbが出力される。
4、 ゲノムストレージの準備ができたら、anvi-pan-genomeプログラムを使ってパンゲノム解析を実行する。
anvi-pan-genome -g PROCHLORO-GENOMES.db -n PROJECT1 -T 40
ディレクトリ PROJECT1/ができ、中にパンゲノムデータベースPROJECT1-PAN.dbなどが出力される。
5、結果を視覚化する。メタゲノムのanvi-interactiveに似たコマンドanvi-display-panを実行する(パンゲノム用の微調整が入っている)。
anvi-display-pan -p PROJECT1/PROJECT1-PAN.db -g PROCHLORO-GENOMES.db
http://localhost:8080 にアクセスする。
Drawで描画

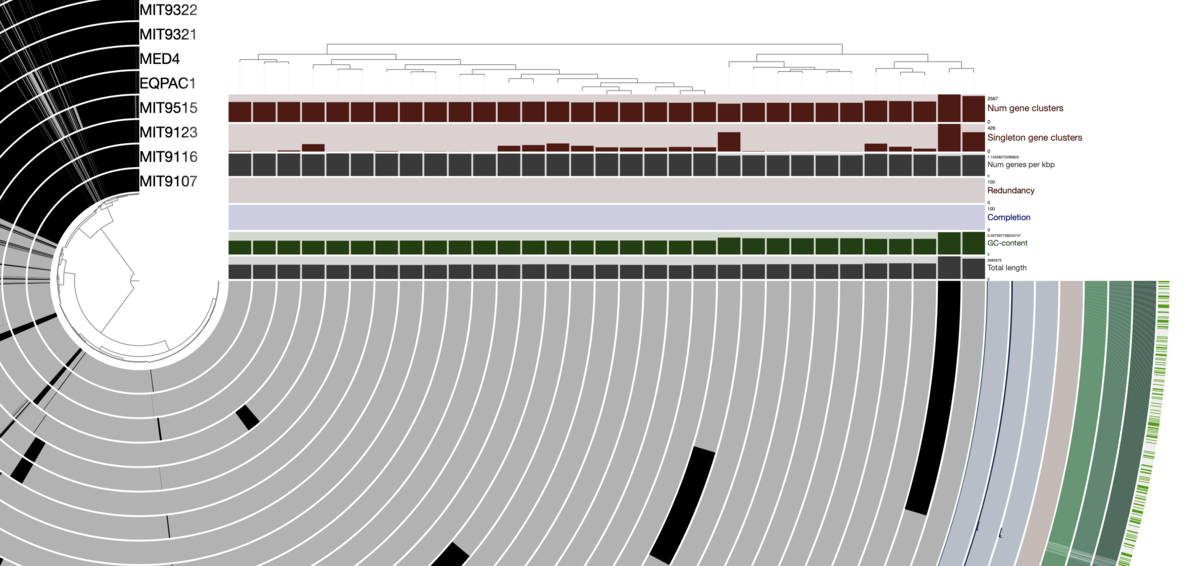
図は左下のボタンやマウスのホイールから自由に拡大縮小できる。環状ゲノムを表した図ではなく、遺伝子の有無を線で表した図である事に注意する。
メニューを隠した。左斜め上の方の、全てのリングが黒くなっている領域がコア遺伝子のクラスタ。左下から右下の方は全てアクセサリ遺伝子。
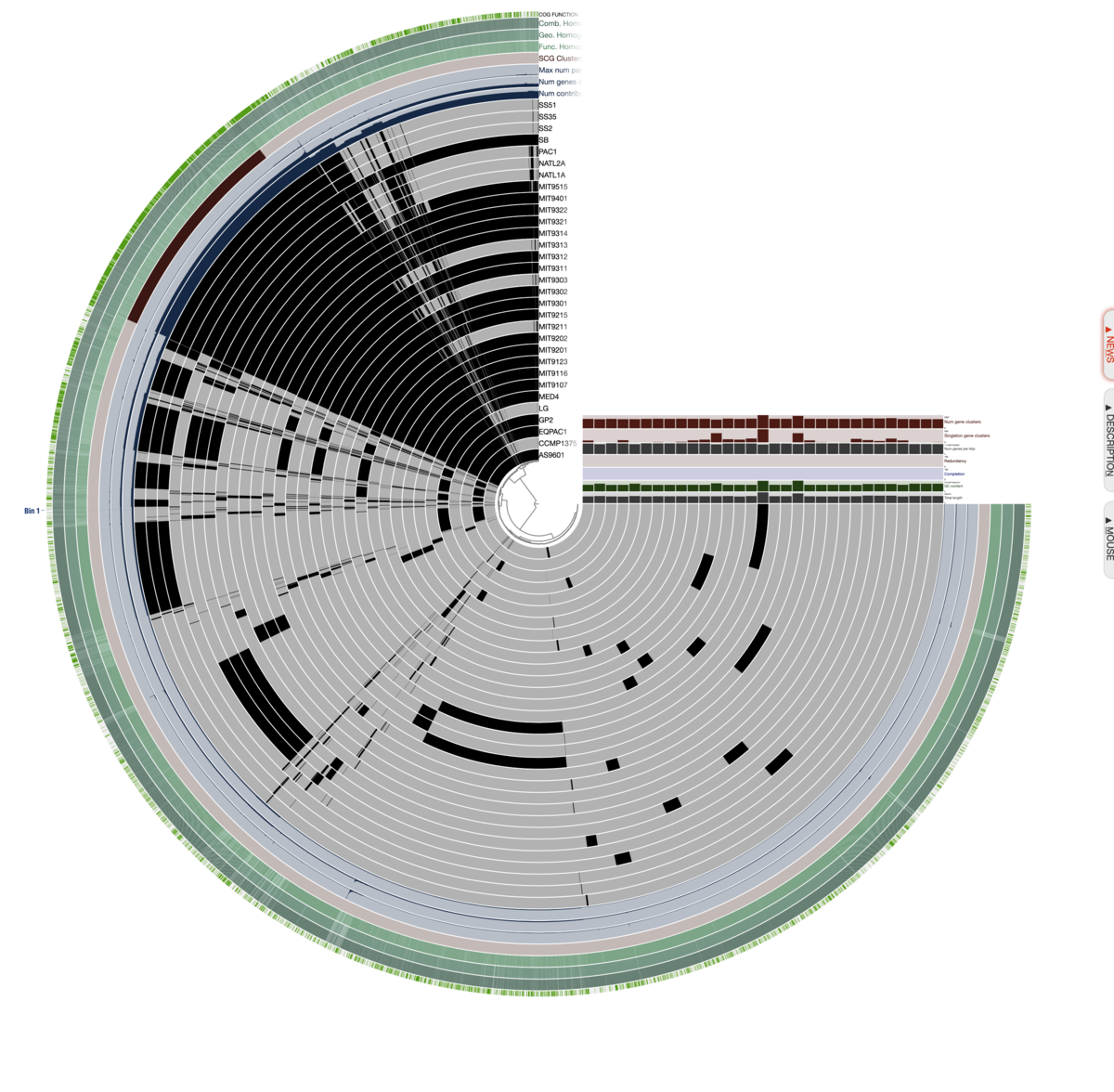
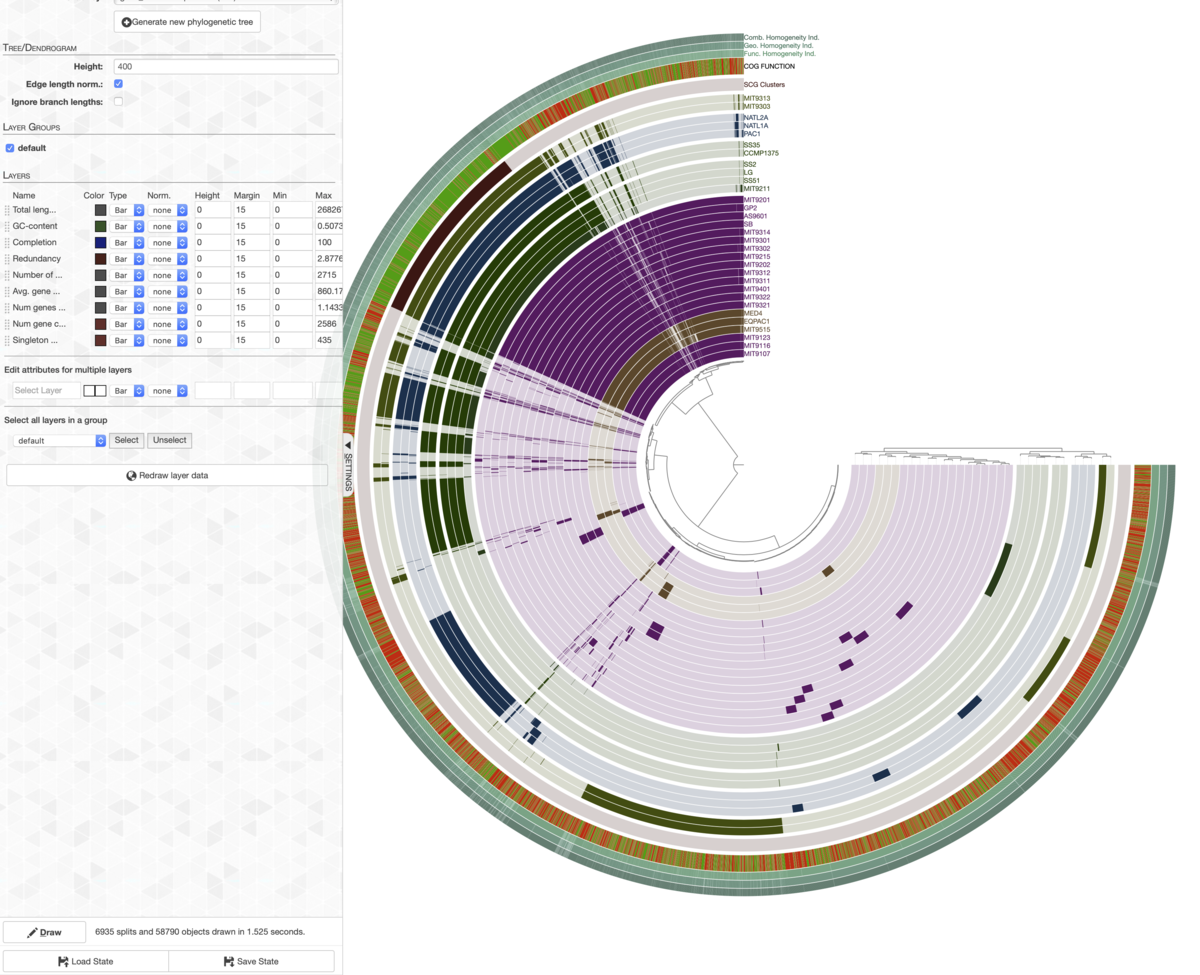
遺伝子クラスタリングの結果に基づいてレイヤーの順番(つまり中心から外周までのリングの順番)を並べ換える。Layerタブ=> Order by => gene_cluster frequenciesを選択。
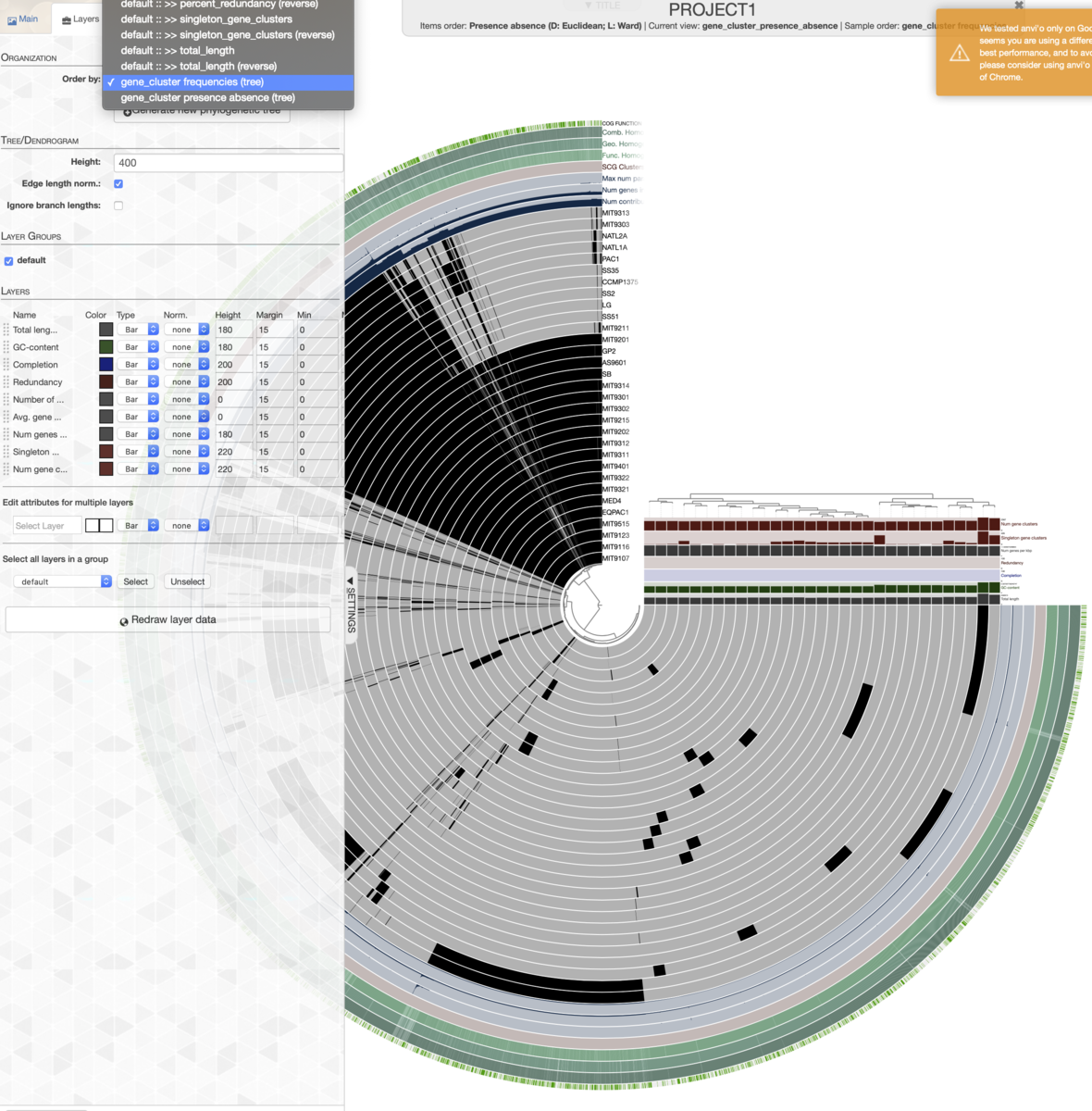
実行すると、クラスタリング結果に基づいてレイヤーの順番が並び替えられ、右上に遺伝子クラスタリング結果のデンドログラムも出現する。
MainタブのItem orderはリング内でのオーダーの指定になる。変更すると、レイヤーの順番は変わらず、1レイヤー内の遺伝子の順番がクラスタリングされる。例えば左斜め上のコア遺伝子クラスタ(真っ黒の部分)が右下に移動したりする。
カスタム設定のプロファイル(JSON)を左下ボタンから設定をexportし、次回起動時にカスタムした設定がデフォルトの状態で視覚化できる。いったんhaltして、以下を実行した。
#自分のprofileをtest1としてexportした場合、--nameをtest1と指定。
anvi-import-state -p PROJECT1/PROJECT1-PAN.db \
--state pan-state.json \
--name default
#再実行。
anvi-display-pan -p PROJECT1/PROJECT1-PAN.db -g PROCHLORO-GENOMES.db
ここではexampleのJSONファイルを読み込ませている。
最初から色などがカスタム設定にした状態で描画された。
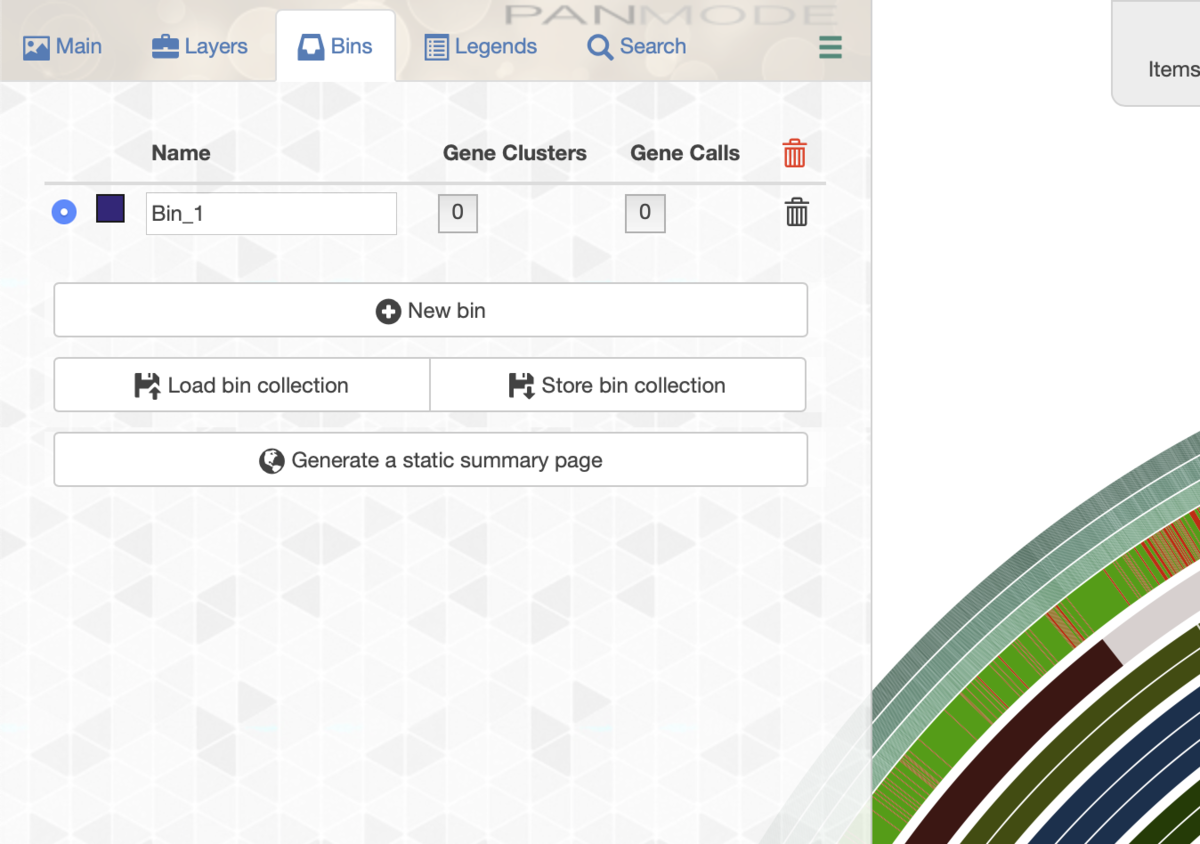
例えば左上の共通して保存されている遺伝子群にコア遺伝子と表記をつけたいとする。その場合、まずBinのタブに移動し、
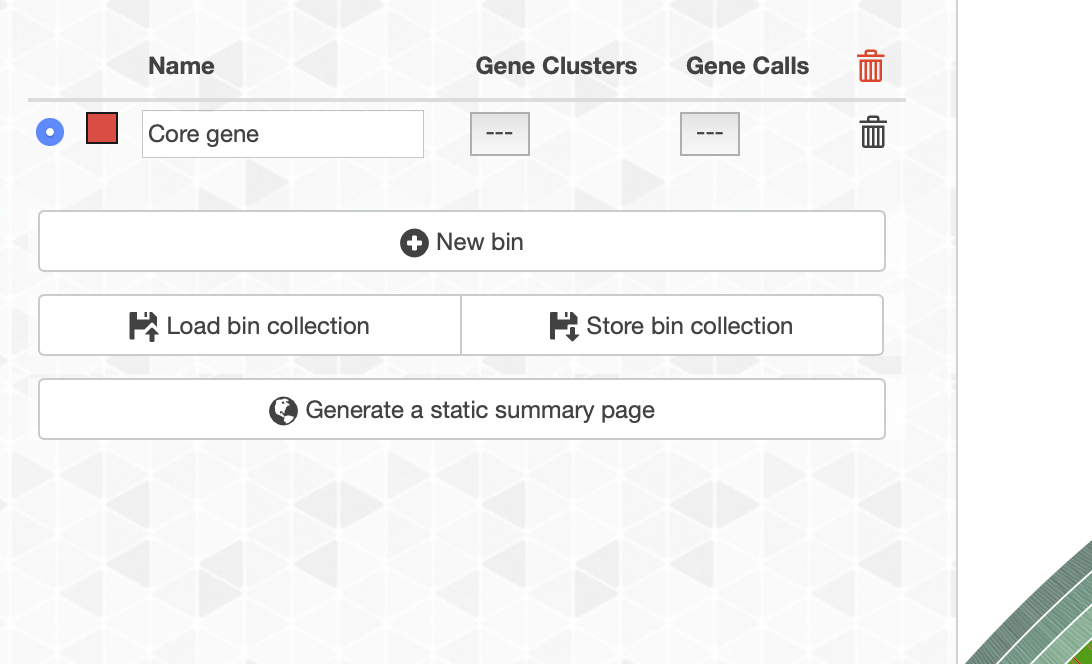
アサインしたい名前をbin_1からcore geneという名前を変更した。色は赤にした。
マウスホイールで少し拡大。中央のデンドログラムのコア遺伝子の枝付近にマウスをホバーすると、下の図のように該当する枝がリアルタイムでハイライト表示されるので。、
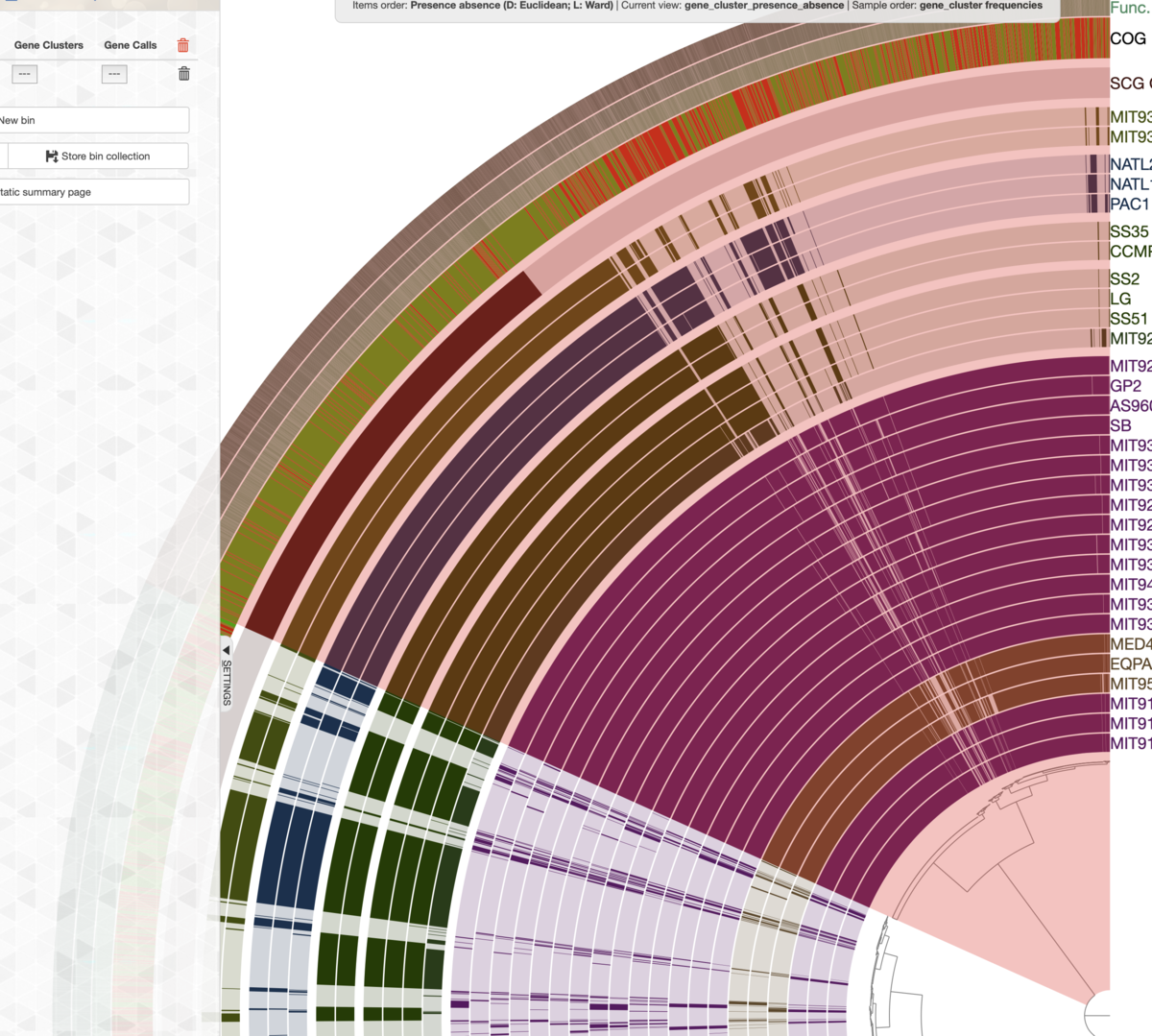
core geneの表記をつけたい枝部分にマウスを合わせる。
ホバーされて色が変わった状態で1回左クリックする。その枝の最外周にCore geneという表記がついた。
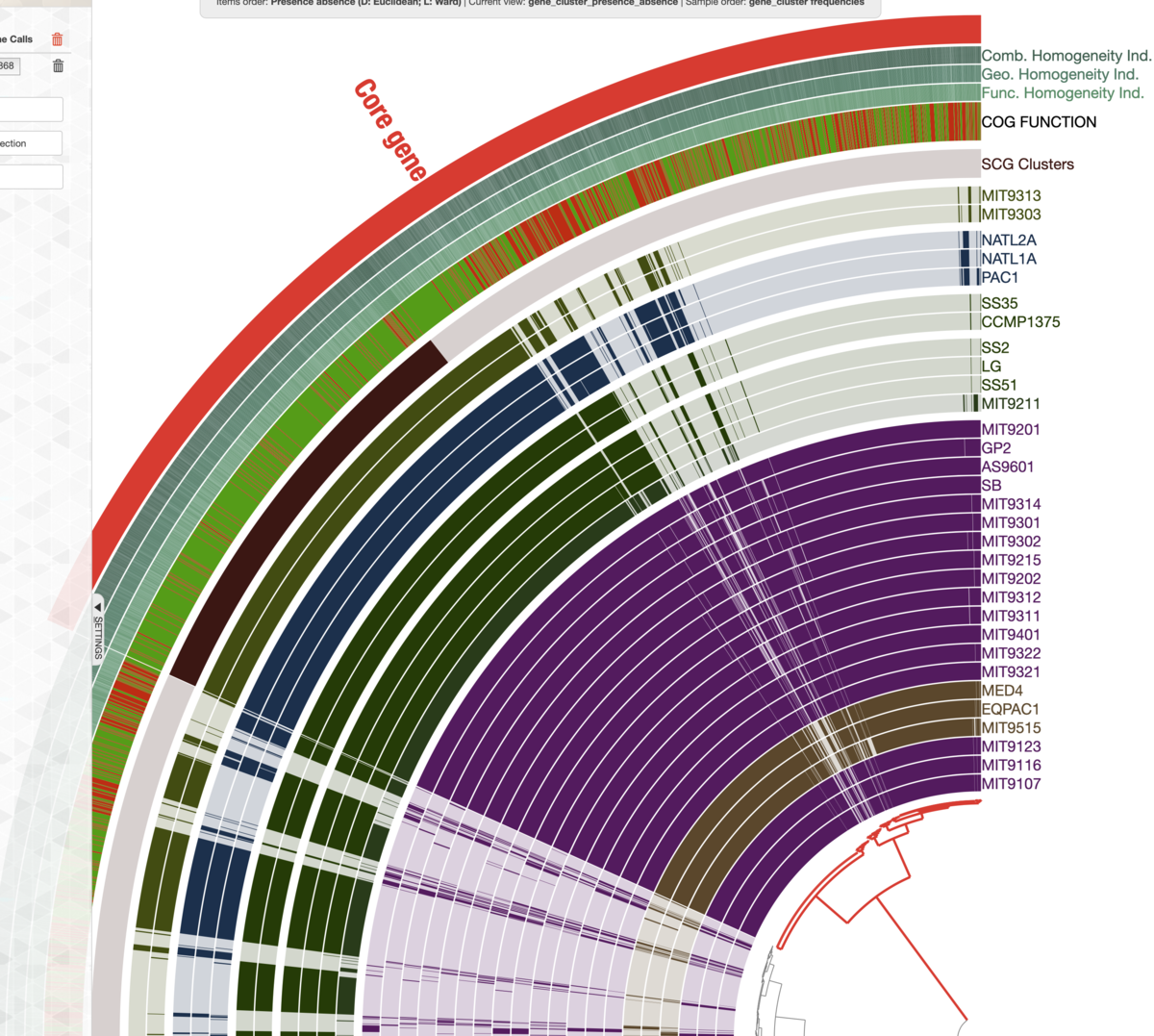
(持たないゲノムまでコア遺伝子に含んでいるので間違ってます)
左上のメニューもプラスをクリック、名前がaccessory gene(色は青)というタグを作成、残りの枝の最外周にaccessory geneという表記をつけた。
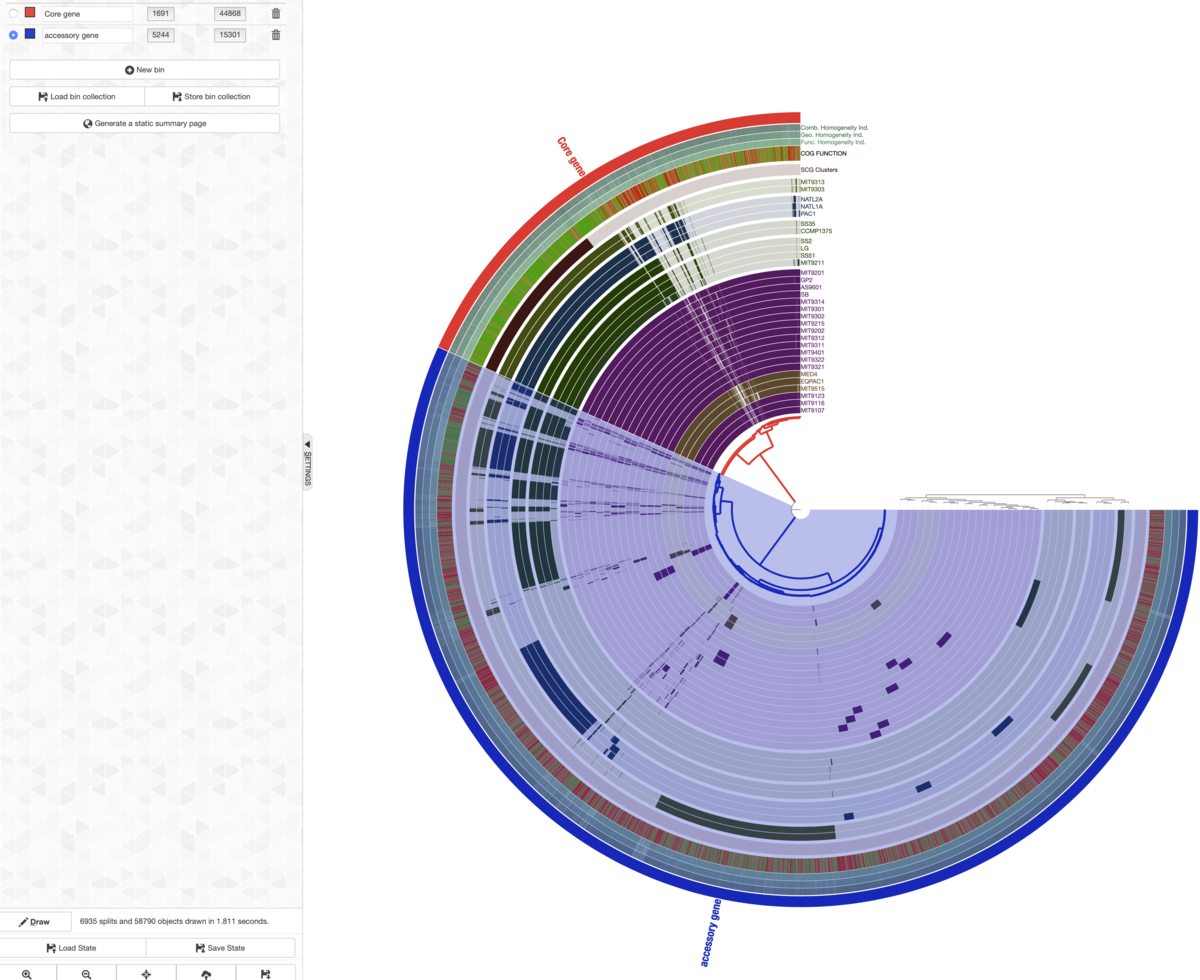
下にaccessory geneの表記をつけた。
大まかな傾向が見られたら、そのうち個別の遺伝子クラスターに興味が出てくる。その場合、関心がある遺伝子クラスターにズームアップし、線の上で右クリックして詳細を調べることができる。

この図では、一番内側の3レイヤーにしかない線を右クリックしている。右端にウィンドウが出現ている。
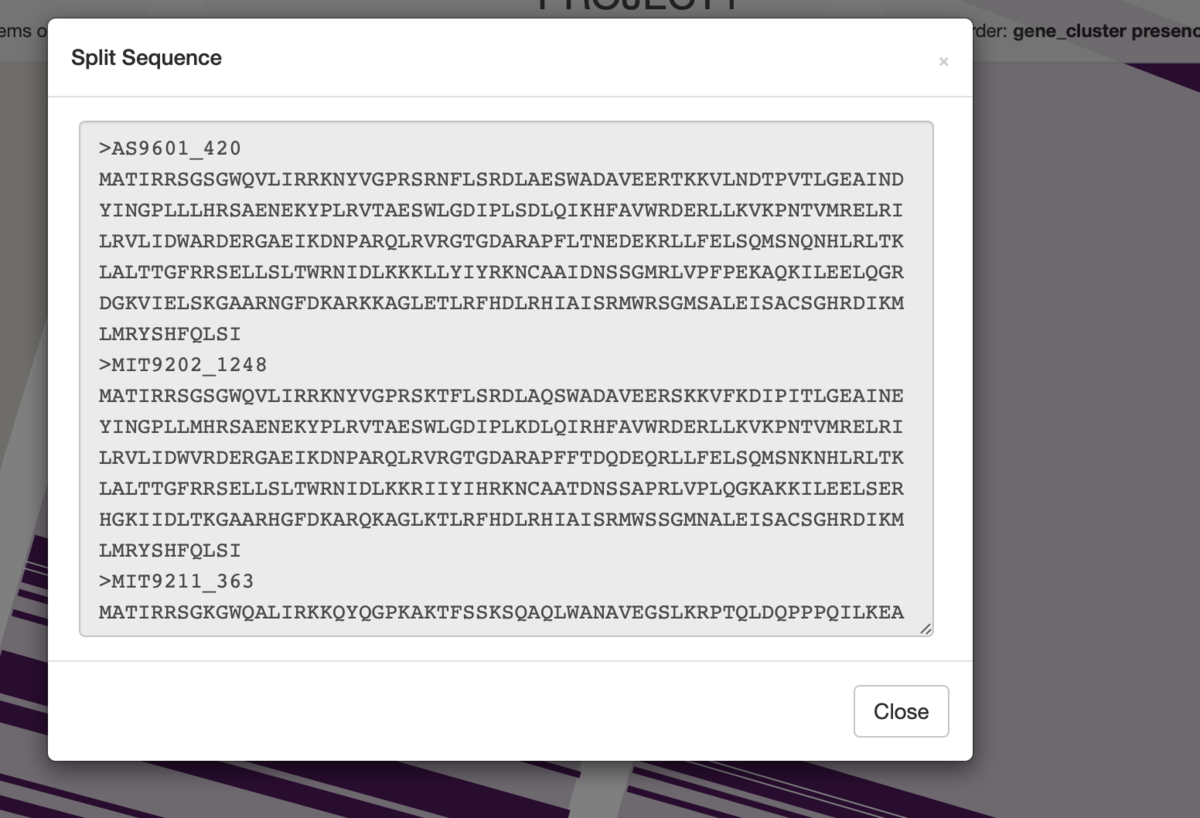
右クリック => ウィンドウのinspect gene clusterを選択すると、新しいウィンドウが生成され。そこに下のように配列が表示される。

線があったゲノム、つまりそのタンパク質が見つかったゲノムだけアミノ酸配列が表示されている。ここでは一番上の3つになる。バッググラウンドの色はリングの色をそのまま反映している。順番も元の通り(上の例だと一番上が円の一番内側)。
右クリックからアミノ酸配列をコピーしたりもできる。
コア遺伝子、アクセサリ遺伝子などに分けて描画する。いったんhaltして、anvi-splitを実行する。
anvi-display-pan -p PROJECT1/PROJECT1-PAN.db -g PROCHLORO-GENOMES.db
ランが終わったら、 出力ディレクトリのコア遺伝子やアクセサリ遺伝子の.dbを指定して描画する。
2020 6/23
52 genome

引用
Anvi'o: an advanced analysis and visualization platform for 'omics data
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO
PeerJ. 2015 Oct 8;3:e1319
関連
作画の参考にした論文
https://www.biorxiv.org/content/10.1101/170639v1.full.pdf