2019 5/7 インストール追記、6/16 パラメータ追記、6/16 upしたdocker イメージのエラー修正、6/18 link追加
2021 4/29 インストール追記、5/18 インストール追記 (condaによるpplacerの導入)、5/27 タイトル変更、5/29, 6/30 compareコマンド追記
2022/06/14 ツイート追記
メタゲノム研究により、シーケンシングされ、ドラフト品質ゲノムが解読される微生物ゲノムの数は毎年急速に拡大している。大きなゲノムセットを包括的に比較するための迅速なアルゴリズムが開発されているが、ドラフト品質のゲノムでは正確ではない。ここでは、不正確だがゲノム距離の迅速かつ推定、および、正確だが遅いANIの計算を順番に適用することによってペアワイズゲノム比較の計算時間を短縮するプログラムdRepを提示する。 dRepは、以前に開発されたアルゴリズムに対してベンチマークされた場合、パーフェクトなrecallとprecisionを維持しながら28倍の速度向上を実現する。我々(著者ら)は、timeseriesデータセットからのゲノムリカバリーでdRepを実証する。時系列の各メタゲノムデータセットは別々にアセンブリされ、それらから、dRepを用いて同一のゲノムグループを同定した。この手順による結果は、時系列データのco-assemblyでリカバリーされたゲノムセットと比べ、はるかにクオリティの高いゲノムリカバリーを達成していた。
drep documentation
https://drep.readthedocs.io/en/latest/overview.html
マニュアルより転載。
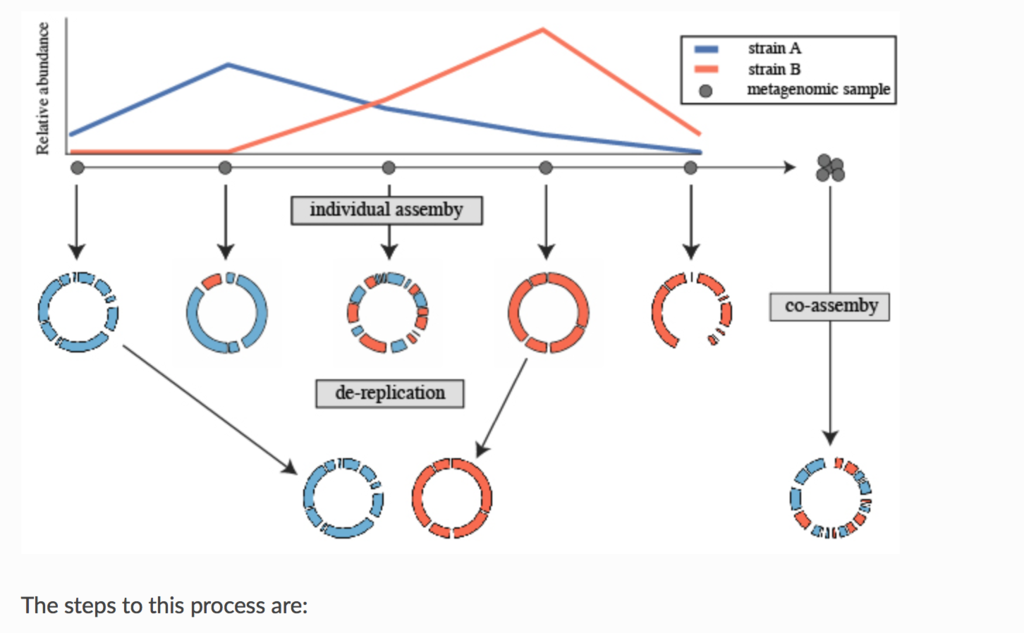
論文のfull textにはアクセスできなかったので分からないが、Prepinrt*1では、よく使われる生後1ヶ月の未熟児の腸内細菌叢 (pubmed)の時系列メタゲノムシーケンシングデータセット(SRA link)を用いて、co-aassemblyと、個別アセンブリ+ dRep(によるde-replication)のアセンブリを比較している。Prepirntの実験結果は、個別アセンブリ+ dRepの方がco-assemblyとその後のanvi’o を使ったマニュアルキュレーションより良好な結果を示している(下の図)。

Prepinrt(*1)のFigure2を転載。
Documentより
Dereplicationとは、ゲノムセットの中から「同じ」ゲノム群を特定し、それぞれのセットから「最適」なゲノムを特定するプロセス。どの程度の類似性があれば「同一」とみなされるか、またどのように「最適」なゲノムを選択するかは、研究に応じて調整することができる。
2024/03/01
Just released version 3.5.0 of dRep (https://t.co/1toeEoNJUx), which now allows skani (https://t.co/xy5H8WMkKx) to be used as a comparison algorithm! (as well as removing a few dependencies that made installation annoying in some cases).
— Matt Olm (@MattagenOlmics) 2024年3月1日
2022/06/14
Just pushed updates to inStrain and dRep: inStrain v1.6.0 provides the ability to pull SNVs from bam files that do not have SNVs called to generate complete frequency tables (https://t.co/gmHZfW3XT7) and dRep v3.3.0 makes fastANI and 95% ANI the default (https://t.co/wyt7u9mH8N)
— Matt Olm (@MattagenOlmics) 2022年6月13日
インストール
依存
- python3(drep -> python >=3.6,<3.7.0a0)
- Mash is used to rapidly compare all genomes in a pair-wise manner(紹介)
- MUMmer is used to perform more actuate comparisons between genomes which are shown to be similar with Mash(紹介)
Optional
- CheckM is used to determine the contamination and completeness of genomes (used during de-replication)(紹介)
- gANI (aka ANIcalculator) is an optional alternative to MUMmer
- Prodigal is a dependency of both checkM and gANI(紹介)
- NSimScan
Accessory
- Centrifuge can be used to perform rough taxonomic assignment of bins(紹介)
参考
checkmはpython2.7系のコード(python >= 2.7 and < 3.0)、他はpython3に移行しているので、pyenvでpythonのバージョンを管理しているなら、pyenvで両方をglobalにして使う必要がある。
checkmは、インストール後、データベースをダウンロードしてパスを指定しておく必要がある(checkmの導入)。
#ANIcalculator
wget https://ani.jgi-psf.org/download_files/ANIcalculator_v1.tgz
tar -zxvf ANIcalculator_v1.tgz
cd ANIcalculator_v1
cp ANIcalculator /usr/local/bin/
git clone https://github.com/abadona/qsimscan.git
cd qsimscan/
make -j 8
#パスを通しておく
#centrifuge
git clone https://github.com/infphilo/centrifuge
cd centrifuge
make -j 8
sudo make install prefix=/usr/local
#fastANI(binary)
wget https://github.com/ParBLiSS/FastANI/releases/download/v1.33/fastANI-Linux64-v1.33.zip
unzip
cp fastANI /usr/local/bin/
本体 Github
#Bioconda (link)
conda install -c bioconda drep
#pip
pip install drep
#pip & conda (2021 4/29)
mamba create -n drep -y python=3.7
conda activate drep
pip install drep
pip install checkm-genome
mamba install -c bioconda -y mash
mamba install -c bioconda -y mummer
mamba install -c bioconda -y fastANI
mamba install -c bioconda -y prodigal
mamba install -c bioconda -y pplacer
#インストール確認
dRep bonus output_directory --check_dependencies
dRep bonus output_directory --check_dependencies
Loading work directory
Checking dependencies
mash.................................... all good (location = /usr/local/bin/mash)
nucmer.................................. all good (location = /opt/conda/bin/nucmer)
checkm.................................. all good (location = /usr/local/bin/checkm)
ANIcalculator........................... all good (location = /gANI/current/ANIcalculator)
prodigal................................ all good (location = /opt/conda/bin/prodigal)
centrifuge.............................. all good (location = /usr/local/bin/centrifuge)
(
O.K (*バージョンアップに伴い現在は必要なものが変わってきています)
> dRep
$ dRep
...::: dRep v3.2.0 :::...
Matt Olm. MIT License. Banfield Lab, UC Berkeley. 2017 (last updated 2020)
See https://drep.readthedocs.io/en/latest/index.html for documentation
Choose one of the operations below for more detailed help.
Example: dRep dereplicate -h
Commands:
compare -> Compare and cluster a set of genomes
dereplicate -> De-replicate a set of genomes
check_dependencies -> Check which dependencies are properly installed
> dRep compare -h
$ dRep compare -h
usage: dRep compare [-p PROCESSORS] [-d] [-h] [-g [GENOMES [GENOMES ...]]]
[--S_algorithm {ANImf,ANIn,fastANI,goANI,gANI}]
[-ms MASH_SKETCH] [--SkipMash] [--SkipSecondary]
[--n_PRESET {normal,tight}] [-pa P_ANI] [-sa S_ANI]
[-nc COV_THRESH] [-cm {total,larger}]
[--clusterAlg {ward,average,complete,median,weighted,centroid,single}]
[--multiround_primary_clustering]
[--primary_chunksize PRIMARY_CHUNKSIZE]
[--greedy_secondary_clustering]
[--run_tertiary_clustering] [--warn_dist WARN_DIST]
[--warn_sim WARN_SIM] [--warn_aln WARN_ALN]
work_directory
positional arguments:
work_directory Directory where data and output are stored
*** USE THE SAME WORK DIRECTORY FOR ALL DREP OPERATIONS ***
SYSTEM PARAMETERS:
-p PROCESSORS, --processors PROCESSORS
threads (default: 6)
-d, --debug make extra debugging output (default: False)
-h, --help show this help message and exit
GENOME INPUT:
-g [GENOMES [GENOMES ...]], --genomes [GENOMES [GENOMES ...]]
genomes to filter in .fasta format. Not necessary if
Bdb or Wdb already exist. Can also input a text file
with paths to genomes, which results in fewer OS
issues than wildcard expansion (default: None)
GENOME COMPARISON OPTIONS:
--S_algorithm {ANImf,ANIn,fastANI,goANI,gANI}
Algorithm for secondary clustering comaprisons:
fastANI = Kmer-based approach; very fast
ANImf = (DEFAULT) Align whole genomes with nucmer; filter alignment; compare aligned regions
ANIn = Align whole genomes with nucmer; compare aligned regions
gANI = Identify and align ORFs; compare aligned ORFS
goANI = Open source version of gANI; requires nsmimscan
(default: ANImf)
-ms MASH_SKETCH, --MASH_sketch MASH_SKETCH
MASH sketch size (default: 1000)
--SkipMash Skip MASH clustering, just do secondary clustering on
all genomes (default: False)
--SkipSecondary Skip secondary clustering, just perform MASH
clustering (default: False)
--n_PRESET {normal,tight}
Presets to pass to nucmer
tight = only align highly conserved regions
normal = default ANIn parameters (default: normal)
GENOME CLUSTERING OPTIONS:
-pa P_ANI, --P_ani P_ANI
ANI threshold to form primary (MASH) clusters
(default: 0.9)
-sa S_ANI, --S_ani S_ANI
ANI threshold to form secondary clusters (default:
0.99)
-nc COV_THRESH, --cov_thresh COV_THRESH
Minmum level of overlap between genomes when doing
secondary comparisons (default: 0.1)
-cm {total,larger}, --coverage_method {total,larger}
Method to calculate coverage of an alignment
(for ANIn/ANImf only; gANI and fastANI can only do larger method)
total = 2*(aligned length) / (sum of total genome lengths)
larger = max*1
(default: larger)
--clusterAlg {ward,average,complete,median,weighted,centroid,single}
Algorithm used to cluster genomes (passed to
scipy.cluster.hierarchy.linkage (default: average)
GREEDY CLUSTERING OPTIONS
These decrease RAM use and runtime at the expense of a minor loss in accuracy.
Recommended when clustering 5000+ genomes:
--multiround_primary_clustering
Cluster each primary clunk separately and merge at the
end with single linkage. Decreases RAM usage and
increases speed, and the cost of a minor loss in
precision and the inability to plot
primary_clustering_dendrograms. Especially helpful
when clustering 5000+ genomes. Will be done with
single linkage clustering (default: False)
--primary_chunksize PRIMARY_CHUNKSIZE
Impacts multiround_primary_clustering. If you have
more than this many genomes, process them in chunks of
this size. (default: 5000)
--greedy_secondary_clustering
Use a heuristic to avoid pair-wise comparisons when
doing secondary clustering. Will be done with single
linkage clustering. Only works for fastANI S_algorithm
option at the moment (default: False)
--run_tertiary_clustering
Run an additional round of clustering on the final
genome set. This is especially useful when greedy
clustering is performed and/or to handle cases where
similar genomes end up in different primary clusters.
Only works with dereplicate, not compare. (default:
False)
WARNINGS:
--warn_dist WARN_DIST
How far from the threshold to throw cluster warnings
(default: 0.25)
--warn_sim WARN_SIM Similarity threshold for warnings between dereplicated
genomes (default: 0.98)
--warn_aln WARN_ALN Minimum aligned fraction for warnings between
dereplicated genomes (ANIn) (default: 0.25)
Example: dRep compare output_dir/ -g /path/to/genomes/*.fasta
> dRep dereplicate -h
$ dRep dereplicate -h
usage: dRep dereplicate [-p PROCESSORS] [-d] [-h] [-g [GENOMES [GENOMES ...]]]
[-l LENGTH] [-comp COMPLETENESS] [-con CONTAMINATION]
[--ignoreGenomeQuality] [--genomeInfo GENOMEINFO]
[--checkM_method {lineage_wf,taxonomy_wf}]
[--set_recursion SET_RECURSION]
[--checkm_group_size CHECKM_GROUP_SIZE]
[--S_algorithm {goANI,gANI,ANIn,fastANI,ANImf}]
[-ms MASH_SKETCH] [--SkipMash] [--SkipSecondary]
[--n_PRESET {normal,tight}] [-pa P_ANI] [-sa S_ANI]
[-nc COV_THRESH] [-cm {total,larger}]
[--clusterAlg {median,centroid,ward,complete,single,average,weighted}]
[--multiround_primary_clustering]
[--primary_chunksize PRIMARY_CHUNKSIZE]
[--greedy_secondary_clustering]
[--run_tertiary_clustering]
[-comW COMPLETENESS_WEIGHT]
[-conW CONTAMINATION_WEIGHT]
[-strW STRAIN_HETEROGENEITY_WEIGHT] [-N50W N50_WEIGHT]
[-sizeW SIZE_WEIGHT] [-centW CENTRALITY_WEIGHT]
[-extraW EXTRA_WEIGHT_TABLE] [--warn_dist WARN_DIST]
[--warn_sim WARN_SIM] [--warn_aln WARN_ALN]
work_directory
positional arguments:
work_directory Directory where data and output are stored
*** USE THE SAME WORK DIRECTORY FOR ALL DREP OPERATIONS ***
SYSTEM PARAMETERS:
-p PROCESSORS, --processors PROCESSORS
threads (default: 6)
-d, --debug make extra debugging output (default: False)
-h, --help show this help message and exit
GENOME INPUT:
-g [GENOMES [GENOMES ...]], --genomes [GENOMES [GENOMES ...]]
genomes to filter in .fasta format. Not necessary if
Bdb or Wdb already exist. Can also input a text file
with paths to genomes, which results in fewer OS
issues than wildcard expansion (default: None)
GENOME FILTERING OPTIONS:
-l LENGTH, --length LENGTH
Minimum genome length (default: 50000)
-comp COMPLETENESS, --completeness COMPLETENESS
Minumum genome completeness (default: 75)
-con CONTAMINATION, --contamination CONTAMINATION
Maximum genome contamination (default: 25)
GENOME QUALITY ASSESSMENT OPTIONS:
--ignoreGenomeQuality
Don't run checkM or do any quality filtering. NOT
RECOMMENDED! This is useful for use with
bacteriophages or eukaryotes or things where checkM
scoring does not work. Will only choose genomes based
on length and N50 (default: False)
--genomeInfo GENOMEINFO
location of .csv file containing quality information
on the genomes. Must contain: ["genome"(basename of
.fasta file of that genome), "completeness"(0-100
value for completeness of the genome),
"contamination"(0-100 value of the contamination of
the genome)] (default: None)
--checkM_method {lineage_wf,taxonomy_wf}
Either lineage_wf (more accurate) or taxonomy_wf
(faster) (default: lineage_wf)
--set_recursion SET_RECURSION
Increases the python recursion limit. NOT RECOMMENDED
unless checkM is crashing due to recursion issues.
Recommended to set to 2000 if needed, but setting this
could crash python (default: 0)
--checkm_group_size CHECKM_GROUP_SIZE
The number of genomes passed to checkM at a time.
Increasing this increases RAM but makes checkM faster
(default: 2000)
GENOME COMPARISON OPTIONS:
--S_algorithm {goANI,gANI,ANIn,fastANI,ANImf}
Algorithm for secondary clustering comaprisons:
fastANI = Kmer-based approach; very fast
ANImf = (DEFAULT) Align whole genomes with nucmer; filter alignment; compare aligned regions
ANIn = Align whole genomes with nucmer; compare aligned regions
gANI = Identify and align ORFs; compare aligned ORFS
goANI = Open source version of gANI; requires nsmimscan
(default: ANImf)
-ms MASH_SKETCH, --MASH_sketch MASH_SKETCH
MASH sketch size (default: 1000)
--SkipMash Skip MASH clustering, just do secondary clustering on
all genomes (default: False)
--SkipSecondary Skip secondary clustering, just perform MASH
clustering (default: False)
--n_PRESET {normal,tight}
Presets to pass to nucmer
tight = only align highly conserved regions
normal = default ANIn parameters (default: normal)
GENOME CLUSTERING OPTIONS:
-pa P_ANI, --P_ani P_ANI
ANI threshold to form primary (MASH) clusters
(default: 0.9)
-sa S_ANI, --S_ani S_ANI
ANI threshold to form secondary clusters (default:
0.99)
-nc COV_THRESH, --cov_thresh COV_THRESH
Minmum level of overlap between genomes when doing
secondary comparisons (default: 0.1)
-cm {total,larger}, --coverage_method {total,larger}
Method to calculate coverage of an alignment
(for ANIn/ANImf only; gANI and fastANI can only do larger method)
total = 2*(aligned length) / (sum of total genome lengths)
larger = max*2
(default: larger)
--clusterAlg {median,centroid,ward,complete,single,average,weighted}
Algorithm used to cluster genomes (passed to
scipy.cluster.hierarchy.linkage (default: average)
GREEDY CLUSTERING OPTIONS
These decrease RAM use and runtime at the expense of a minor loss in accuracy.
Recommended when clustering 5000+ genomes:
--multiround_primary_clustering
Cluster each primary clunk separately and merge at the
end with single linkage. Decreases RAM usage and
increases speed, and the cost of a minor loss in
precision and the inability to plot
primary_clustering_dendrograms. Especially helpful
when clustering 5000+ genomes. Will be done with
single linkage clustering (default: False)
--primary_chunksize PRIMARY_CHUNKSIZE
Impacts multiround_primary_clustering. If you have
more than this many genomes, process them in chunks of
this size. (default: 5000)
--greedy_secondary_clustering
Use a heuristic to avoid pair-wise comparisons when
doing secondary clustering. Will be done with single
linkage clustering. Only works for fastANI S_algorithm
option at the moment (default: False)
--run_tertiary_clustering
Run an additional round of clustering on the final
genome set. This is especially useful when greedy
clustering is performed and/or to handle cases where
similar genomes end up in different primary clusters.
Only works with dereplicate, not compare. (default:
False)
SCORING CRITERIA
Based off of the formula:
A*Completeness - B*Contamination + C*(Contamination * (strain_heterogeneity/100)) + D*log(N50) + E*log(size) + F*(centrality - S_ani)
A = completeness_weight; B = contamination_weight; C = strain_heterogeneity_weight; D = N50_weight; E = size_weight; F = cent_weight:
-comW COMPLETENESS_WEIGHT, --completeness_weight COMPLETENESS_WEIGHT
completeness weight (default: 1)
-conW CONTAMINATION_WEIGHT, --contamination_weight CONTAMINATION_WEIGHT
contamination weight (default: 5)
-strW STRAIN_HETEROGENEITY_WEIGHT, --strain_heterogeneity_weight STRAIN_HETEROGENEITY_WEIGHT
strain heterogeneity weight (default: 1)
-N50W N50_WEIGHT, --N50_weight N50_WEIGHT
weight of log(genome N50) (default: 0.5)
-sizeW SIZE_WEIGHT, --size_weight SIZE_WEIGHT
weight of log(genome size) (default: 0)
-centW CENTRALITY_WEIGHT, --centrality_weight CENTRALITY_WEIGHT
Weight of (centrality - S_ani) (default: 1)
-extraW EXTRA_WEIGHT_TABLE, --extra_weight_table EXTRA_WEIGHT_TABLE
Path to a tab-separated file with two-columns, no
headers, listing genome and extra score to apply to
that genome (default: None)
WARNINGS:
--warn_dist WARN_DIST
How far from the threshold to throw cluster warnings
(default: 0.25)
--warn_sim WARN_SIM Similarity threshold for warnings between dereplicated
genomes (default: 0.98)
--warn_aln WARN_ALN Minimum aligned fraction for warnings between
dereplicated genomes (ANIn) (default: 0.25)
Example: dRep dereplicate output_dir/ -g /path/to/genomes/*.fasta
> dRep compare -h
$ dRep compare -h
usage: dRep compare [-p PROCESSORS] [-d] [-h] [-g [GENOMES [GENOMES ...]]]
[--S_algorithm {gANI,fastANI,ANImf,ANIn,goANI}]
[-ms MASH_SKETCH] [--SkipMash] [--SkipSecondary]
[--n_PRESET {normal,tight}] [-pa P_ANI] [-sa S_ANI]
[-nc COV_THRESH] [-cm {total,larger}]
[--clusterAlg {ward,median,average,single,weighted,complete,centroid}]
[--multiround_primary_clustering]
[--primary_chunksize PRIMARY_CHUNKSIZE]
[--greedy_secondary_clustering]
[--run_tertiary_clustering] [--warn_dist WARN_DIST]
[--warn_sim WARN_SIM] [--warn_aln WARN_ALN]
work_directory
positional arguments:
work_directory Directory where data and output are stored
*** USE THE SAME WORK DIRECTORY FOR ALL DREP OPERATIONS ***
SYSTEM PARAMETERS:
-p PROCESSORS, --processors PROCESSORS
threads (default: 6)
-d, --debug make extra debugging output (default: False)
-h, --help show this help message and exit
GENOME INPUT:
-g [GENOMES [GENOMES ...]], --genomes [GENOMES [GENOMES ...]]
genomes to filter in .fasta format. Not necessary if
Bdb or Wdb already exist. Can also input a text file
with paths to genomes, which results in fewer OS
issues than wildcard expansion (default: None)
GENOME COMPARISON OPTIONS:
--S_algorithm {gANI,fastANI,ANImf,ANIn,goANI}
Algorithm for secondary clustering comaprisons:
fastANI = Kmer-based approach; very fast
ANImf = (DEFAULT) Align whole genomes with nucmer; filter alignment; compare aligned regions
ANIn = Align whole genomes with nucmer; compare aligned regions
gANI = Identify and align ORFs; compare aligned ORFS
goANI = Open source version of gANI; requires nsmimscan
(default: ANImf)
-ms MASH_SKETCH, --MASH_sketch MASH_SKETCH
MASH sketch size (default: 1000)
--SkipMash Skip MASH clustering, just do secondary clustering on
all genomes (default: False)
--SkipSecondary Skip secondary clustering, just perform MASH
clustering (default: False)
--n_PRESET {normal,tight}
Presets to pass to nucmer
tight = only align highly conserved regions
normal = default ANIn parameters (default: normal)
GENOME CLUSTERING OPTIONS:
-pa P_ANI, --P_ani P_ANI
ANI threshold to form primary (MASH) clusters
(default: 0.9)
-sa S_ANI, --S_ani S_ANI
ANI threshold to form secondary clusters (default:
0.99)
-nc COV_THRESH, --cov_thresh COV_THRESH
Minmum level of overlap between genomes when doing
secondary comparisons (default: 0.1)
-cm {total,larger}, --coverage_method {total,larger}
Method to calculate coverage of an alignment
(for ANIn/ANImf only; gANI and fastANI can only do larger method)
total = 2*(aligned length) / (sum of total genome lengths)
larger = max*3
(default: larger)
--clusterAlg {ward,median,average,single,weighted,complete,centroid}
Algorithm used to cluster genomes (passed to
scipy.cluster.hierarchy.linkage (default: average)
GREEDY CLUSTERING OPTIONS
These decrease RAM use and runtime at the expense of a minor loss in accuracy.
Recommended when clustering 5000+ genomes:
--multiround_primary_clustering
Cluster each primary clunk separately and merge at the
end with single linkage. Decreases RAM usage and
increases speed, and the cost of a minor loss in
precision and the inability to plot
primary_clustering_dendrograms. Especially helpful
when clustering 5000+ genomes. Will be done with
single linkage clustering (default: False)
--primary_chunksize PRIMARY_CHUNKSIZE
Impacts multiround_primary_clustering. If you have
more than this many genomes, process them in chunks of
this size. (default: 5000)
--greedy_secondary_clustering
Use a heuristic to avoid pair-wise comparisons when
doing secondary clustering. Will be done with single
linkage clustering. Only works for fastANI S_algorithm
option at the moment (default: False)
--run_tertiary_clustering
Run an additional round of clustering on the final
genome set. This is especially useful when greedy
clustering is performed and/or to handle cases where
similar genomes end up in different primary clusters.
Only works with dereplicate, not compare. (default:
False)
WARNINGS:
--warn_dist WARN_DIST
How far from the threshold to throw cluster warnings
(default: 0.25)
--warn_sim WARN_SIM Similarity threshold for warnings between dereplicated
genomes (default: 0.98)
--warn_aln WARN_ALN Minimum aligned fraction for warnings between
dereplicated genomes (ANIn) (default: 0.25)
Example: dRep compare output_dir/ -g /path/to/genomes/*.fasta
導入に手間取ったので、環境構築した後のイメージをpdocker hubにpushしておきます。pullする場合、別のツールのテストもしていたので、少しイメージが大きくなっています。ご注意下さい(注意;古くなっています)。
追記
checkmのstepでエラーが出たのでBug fixしてpushし直しました。
docker pull kazumax/drep
#カレントパスと/shareを共有して立ち上げる。"--rm"をつけるとexitで廃棄
docker run --rm -itv $PWD:/data/ kazumax/drep
#usage:
source ${HOME}/.bash_profile
#check dependency
dRep bonus output_directory --check_dependencies
#2022/03/06 v3.0
docker pull kazumax/drep:3.0
source ${HOME}/.bashrc
dRep check_dependencies
実行方法
1、ゲノムの比較
dRep compare output_directory -g RefSeq/*.fasta
結果は可視化される。
RefSeqのゲノム(ダウンロード)をいくつか選んで比較してみた。
出力ディレクトリ




2、de-replication
dRep dereplicate outout_directory -g path/to/genomes/*.fasta \
-p 12 -l 50000
- -p threads (default: 6)
- -l Minimum genome length (default: 50000)
- -g genomes to cluster in .fasta format (default: None)
- --checkM_method {taxonomy_wf, lineage_wf} Either lineage_wf (more accurate) or taxonomy_wf (faster) (default: lineage_wf)
結果は可視化される。
出力ディレクトリ

figures




出力の詳細はmanual参照。
2021 5/18
1次クラスタリングでは90%でラフにクラスタリング(図の黒い破線)、2次クラスタリングではgANIを使って95%閾値でde-replication(mOTUsの取得)。5000bp以上の配列を対象とする。
dRep dereplicate outdir -g maxbin2/*fa \
-p 40 -l 5000 -pa 0.90 -sa 0.95 -comp 75 -con 25 -nc 0.1
- -pa ANI threshold to form primary (MASH) clusters (default: 0.9)
- -sa ANI threshold to form secondary clusters (default: 0.99)
- -nc Minmum level of overlap between genomes when doing secondary comparisons (default: 0.1)
- -l Minimum genome length (default: 50000)
- -comp Minumum genome completeness (default: 75)
- -con Maximum genome contamination (default: 25)
compareコマンドを使ってbinned.fastaと既知ゲノム配列を比較する。
dRep compare outdir -g genome/*fna
メモ
- 例えば、AがBに似ていて、BがCに似ていて、AとCが似ていないこのようなケースが存在すると仮定すると、 ANIがしきい値より大きいゲノムペアが異なるクラスタに入ってしまう可能性があるとされる(link)。
=> シングルモード(-clusterAlg single)で実行する
- バクテリア以外の存在(ファージなど)が予想されて、checkMで評価できない場合、 -ignoreGenomeQualityというフラグを立てて、品質フィルタリングやゲノム選択時の完全性・汚染性の使用をオフにすることができる。
- Mashは不完全なゲノム間の距離を過小評価し 、同一種のゲノムを複数のゲノムビンに分割してしまうことがあるため、第一段階のall versus all比較ではMashを使い、それから、精度が高い方法で2回目のクラスタリングを行う。
de-replicationは例えば以下の有名な論文で使用されています。パラメータも記載されています。
https://www.nature.com/articles/s41467-018-03317-6
こちらのプレプリントにもパラメータ例があります。
https://www.biorxiv.org/content/10.1101/2021.04.02.438222v1.full.pdf
Paleofecesをターゲットにした
Reconstruction of ancient microbial genomes from the human gut
https://www.nature.com/articles/s41586-021-03532-0
の論文でも使用されています。
引用
dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replicatio
Olm MR, Brown CT, Brooks B, Banfield JF
ISME J. 2017 Dec;11(12):2864-2868
*1
Preprint
https://www.biorxiv.org/content/biorxiv/early/2017/02/13/108142.full.pdf
関連ツール
BMScan
Mash
Checkm
Centrifuge
2022/02/28
checkM2のランタイムを入れてもこちらの方がdRepよりずっと高速。ANI99以上にも対応している点も良い。